Gene
KWMTBOMO09660
Pre Gene Modal
BGIBMGA012871
Annotation
PREDICTED:_apyrase_[Amyelois_transitella]
Full name
Apyrase
Alternative Name
ATP-diphosphatase
ATP-diphosphohydrolase
Adenosine diphosphatase
ATP-diphosphohydrolase
Adenosine diphosphatase
Location in the cell
PlasmaMembrane Reliability : 1.682
Sequence
CDS
ATGTGGAAACTAGTTTTGTATCTTGTAGTTCTCGGCACAGTTTTGGGACAATCTGACAATGGGCTTTTCGAATTAAACATCATTCACTATAACGATTTCCACGCACACTTCGACGAGATTAGTCCACGAGGAACGATCTGTAATCCCACCGAAGACCCATGCATCGGTGGTTTTGCTAGGCTGTATACAGCCATTATGGAGATTTTTGAAGCCGAACCAGATTCGCTACTTCTTAACGGAGGGGACACGTTCCAAGGAACAATCTGGTACAACTTCTTGAGATGGAATGTCACTCAACATTTTATGAACATGCTGCCACATGATGCACACGTACTCGGAAACCACGAGTTCGATCATGGAGTGGAAGGTCTATTGCCGTATTTGGAACGTTTAAATTCCCCAATGCTAGGTGCTAATGTTAATACAACTTTTGAACCATCTTTGACGCCATTCGTGAAAAATCACGTAATTGTAGAACGAAATGGTAGGAAGATTGGTATCATAGGTGTTTTGTTGCGAGTGTTTTCAGCGCCAATTGGTCAAGTCATCATGGAAGATGAACTTGAGGCTGTAAATCGAGAAGCGGCCATACTAACCGAAGCCGGAGTTGATATTATAATTGTACTCTCTCATATTGGATACACCAGTGATTTAAATCTAGCGGCCCGCGTTGTTCCGGAAGTGGATATCATAGTTGGCGCGCATTCCCATACACTGTTGTTCAACGGAGAAACTCCTAATGGTGACACGGCATTAGGGGAATACCCGACTGTTGTCACACAAGATAATGGACACAGGGTACTGGTGGTTCAAGCGTCTTGTTACACGCGATATTTGGGCGACATAAAACTGTATATCAACGATAGAGGCATTATTGAAAGCTGGGAAGGGCTGCCTGTTTTCTTAGGATCATCTATTGTACAAGATCCTAGGATCCTGGATGAATTAGAGCCGTGGCGTCAGGAGGTGGATGCGATTGGTAAAGAGGTGCTCGGCACTACACTGACGAGTCTCAGCCGAGGATGTACGCGCCGTGAGTGTAACCTTGGCAGCTGGGCGTGTGATGGATTTCTAGAAGAGATGTTGTCACAAGCTACGGGTACAGCGTGGAATGGAGCTCATCTCTGCTTAATAAATTCGGGCGGATTGCGCGTGCAAATAGCACCTGGCGAGGTAACAACTGAGGGACTGCTAATGGCTATACCGTTTGAGAATTACATCCAAGTGTACGACCTAAAGGGTGAATACCTACTCGAGGCGTTAGAGTTTTCTGTGGGTGTGGCACAGACAACACCGGGAGACTTCTCTAGTTCACGAATGTTACAAATAGGAGGGATGCGCAATGTATACAATGCGTCAGCTCCGATCGGTTCCCGTGTGATTTCCGCGACGGTCCGGTGCATAGAGTGCGACGTACCGCAGTATGAGCCCCTCGACGTCACCGCCACCTACAGAGTGGTGACGCAAAACTATATCGGAGATGGAGGCGGCGGTTACACGATGCTGTCGAACAACAGAGAAAACCTCGAGAATTTGGATGTCGACTACGTAGTGTTACAACGTTACATGCGTAGACAACAAACAGTACTGCAAGACCTGGACGGACGCATACAGATTGTGTATTAA
Protein
MWKLVLYLVVLGTVLGQSDNGLFELNIIHYNDFHAHFDEISPRGTICNPTEDPCIGGFARLYTAIMEIFEAEPDSLLLNGGDTFQGTIWYNFLRWNVTQHFMNMLPHDAHVLGNHEFDHGVEGLLPYLERLNSPMLGANVNTTFEPSLTPFVKNHVIVERNGRKIGIIGVLLRVFSAPIGQVIMEDELEAVNREAAILTEAGVDIIIVLSHIGYTSDLNLAARVVPEVDIIVGAHSHTLLFNGETPNGDTALGEYPTVVTQDNGHRVLVVQASCYTRYLGDIKLYINDRGIIESWEGLPVFLGSSIVQDPRILDELEPWRQEVDAIGKEVLGTTLTSLSRGCTRRECNLGSWACDGFLEEMLSQATGTAWNGAHLCLINSGGLRVQIAPGEVTTEGLLMAIPFENYIQVYDLKGEYLLEALEFSVGVAQTTPGDFSSSRMLQIGGMRNVYNASAPIGSRVISATVRCIECDVPQYEPLDVTATYRVVTQNYIGDGGGGYTMLSNNRENLENLDVDYVVLQRYMRRQQTVLQDLDGRIQIVY
Summary
Description
Facilitates hematophagy by inhibiting ADP-dependent platelet aggregation in the host. Shows potential for antithrombotic activity. May reduce probing time by facilitating the speed of locating blood.
Catalytic Activity
a ribonucleoside 5'-triphosphate + 2 H2O = a ribonucleoside 5'-phosphate + 2 H(+) + 2 phosphate
Cofactor
a divalent metal cation
Similarity
Belongs to the 5'-nucleotidase family.
Keywords
Allergen
ATP-binding
Direct protein sequencing
Hydrolase
Metal-binding
Nucleotide-binding
Secreted
Signal
Feature
chain Apyrase
Uniprot
H9JTK9
A0A2A4JWR7
A0A2A4JW12
A0A437B966
A0A212FMH9
A0A194Q670
+ More
A0A2A4JX59 A0A194R1G7 A0A2A4JW25 A0A3S2M1V1 A0A2A4JVW4 A0A194R505 H9JL90 A0A2H1WZX1 A0A194R6M3 B0WJ00 A0A194PLJ4 A0A194PMT9 A0A1Q3FIZ4 A0A182U110 A0A2C9GRQ1 A0A182GQ46 A0A1Q3FN26 A0A1C7CZG9 A0A067RQX4 A0A182VC59 A0A1S4KDB8 Q7QIZ1 Q16QA5 A0A182HLB4 Q8T5H5 Q17NS8 A0A182L9P8 A0A182SCB1 Q8MU74 A0A182WSG4 A0A182V0W0 A0A182R636 A0A182VZA6 A0A182MGY7 A0A2J7RI11 A0A182TN56 A0A336LSN5 A0A1L8ECN7 A0A1I8PB47 A0A336LWJ0 A0A1Y1N7H1 A0A182IVB6 A0A084VT67 A0A182YKS7 A0A1I8NGR4 A0A182R0R0 A0A182K4G0 A0A182YAL5 V5G6C5 B4NEB2 U5EW72 A0A182GQ47 T1PD85 A0A336KUY8 Q16UA0 W5JVI8 A0A182M997 Q16QA4 A0A084VT66 A0A034WIT9 Q9U9I6 A0A034WFB4 B0XGC2 A0A2M4AV02 A0A0K8WAX8 A0A0K8VPE5 A0A182GQ45 W5JSD1 B4MAL8 W8BUB4 A0A182NS57 A0A1L8DPP0 A0A1L8DPP4 A0A182J0X1 A0A182JMY7 A0A182PGQ3 A0A0Q9X374 B4L649 A0A0R3P4U5 A0A182WX10 A0A0A1WD31 A0A1I8NCS8 B3NVB0 A0A182Q352 A0A1L8DPX0 F1JZ10 B3A0N5 A0A182QET4 A0A182PGQ4 A0A182PCT2 J3JZ21 A0A3G1T1Q5 W5J3U7
A0A2A4JX59 A0A194R1G7 A0A2A4JW25 A0A3S2M1V1 A0A2A4JVW4 A0A194R505 H9JL90 A0A2H1WZX1 A0A194R6M3 B0WJ00 A0A194PLJ4 A0A194PMT9 A0A1Q3FIZ4 A0A182U110 A0A2C9GRQ1 A0A182GQ46 A0A1Q3FN26 A0A1C7CZG9 A0A067RQX4 A0A182VC59 A0A1S4KDB8 Q7QIZ1 Q16QA5 A0A182HLB4 Q8T5H5 Q17NS8 A0A182L9P8 A0A182SCB1 Q8MU74 A0A182WSG4 A0A182V0W0 A0A182R636 A0A182VZA6 A0A182MGY7 A0A2J7RI11 A0A182TN56 A0A336LSN5 A0A1L8ECN7 A0A1I8PB47 A0A336LWJ0 A0A1Y1N7H1 A0A182IVB6 A0A084VT67 A0A182YKS7 A0A1I8NGR4 A0A182R0R0 A0A182K4G0 A0A182YAL5 V5G6C5 B4NEB2 U5EW72 A0A182GQ47 T1PD85 A0A336KUY8 Q16UA0 W5JVI8 A0A182M997 Q16QA4 A0A084VT66 A0A034WIT9 Q9U9I6 A0A034WFB4 B0XGC2 A0A2M4AV02 A0A0K8WAX8 A0A0K8VPE5 A0A182GQ45 W5JSD1 B4MAL8 W8BUB4 A0A182NS57 A0A1L8DPP0 A0A1L8DPP4 A0A182J0X1 A0A182JMY7 A0A182PGQ3 A0A0Q9X374 B4L649 A0A0R3P4U5 A0A182WX10 A0A0A1WD31 A0A1I8NCS8 B3NVB0 A0A182Q352 A0A1L8DPX0 F1JZ10 B3A0N5 A0A182QET4 A0A182PGQ4 A0A182PCT2 J3JZ21 A0A3G1T1Q5 W5J3U7
EC Number
3.6.1.5
Pubmed
EMBL
BABH01003658
BABH01003659
NWSH01000463
PCG76226.1
PCG76227.1
RSAL01000115
+ More
RVE46873.1 AGBW02007651 OWR54937.1 KQ459582 KPI98905.1 PCG76224.1 KQ460878 KPJ11542.1 PCG76225.1 RSAL01000067 RVE49304.1 NWSH01000580 PCG75530.1 KQ460685 KPJ12888.1 BABH01011796 ODYU01012308 SOQ58557.1 KPJ12890.1 DS231954 EDS28861.1 KQ459600 KPI94306.1 KPI94308.1 GFDL01007465 JAV27580.1 APCN01000169 JXUM01079657 JXUM01079658 JXUM01079659 KQ563139 KXJ74403.1 GFDL01006108 JAV28937.1 KK852475 KDR23050.1 AAAB01008807 EAA04249.4 CH477755 EAT36583.1 APCN01000890 AJ439398 AAAB01008987 CAD28126.1 EAA00943.2 CH477196 EAT48396.1 AJ441131 CAD29633.1 AXCM01018154 NEVH01003506 PNF40456.1 UFQT01000158 SSX21032.1 GFDG01002404 JAV16395.1 SSX21033.1 GEZM01010744 JAV93862.1 ATLV01016253 KE525074 KFB41161.1 AXCN02000433 GALX01002836 JAB65630.1 CH964239 EDW82081.2 GANO01000692 JAB59179.1 JXUM01079660 KXJ74404.1 KA646644 AFP61273.1 UFQS01001030 UFQT01001030 SSX08629.1 SSX28545.1 CH477627 EAT38116.1 ADMH02000310 ETN67025.1 AXCM01013925 EAT36584.1 KFB41160.1 GAKP01004725 JAC54227.1 AF169229 AAD49730.1 GAKP01004726 JAC54226.1 DS233012 EDS27504.1 GGFK01011279 MBW44600.1 GDHF01004097 JAI48217.1 GDHF01011563 JAI40751.1 JXUM01079656 KXJ74402.1 ETN67026.1 CH940655 EDW66277.1 GAMC01013841 JAB92714.1 GFDF01005651 JAV08433.1 GFDF01005652 JAV08432.1 KRF99569.1 CH933812 EDW05845.1 CH379064 KRT06403.1 GBXI01017413 JAC96878.1 CH954180 EDV46098.2 AXCN02000266 GFDF01005657 JAV08427.1 JF308210 ADX78255.1 BT128502 AEE63459.1 MG770323 AXY94925.1 ADMH02002183 ETN57988.1
RVE46873.1 AGBW02007651 OWR54937.1 KQ459582 KPI98905.1 PCG76224.1 KQ460878 KPJ11542.1 PCG76225.1 RSAL01000067 RVE49304.1 NWSH01000580 PCG75530.1 KQ460685 KPJ12888.1 BABH01011796 ODYU01012308 SOQ58557.1 KPJ12890.1 DS231954 EDS28861.1 KQ459600 KPI94306.1 KPI94308.1 GFDL01007465 JAV27580.1 APCN01000169 JXUM01079657 JXUM01079658 JXUM01079659 KQ563139 KXJ74403.1 GFDL01006108 JAV28937.1 KK852475 KDR23050.1 AAAB01008807 EAA04249.4 CH477755 EAT36583.1 APCN01000890 AJ439398 AAAB01008987 CAD28126.1 EAA00943.2 CH477196 EAT48396.1 AJ441131 CAD29633.1 AXCM01018154 NEVH01003506 PNF40456.1 UFQT01000158 SSX21032.1 GFDG01002404 JAV16395.1 SSX21033.1 GEZM01010744 JAV93862.1 ATLV01016253 KE525074 KFB41161.1 AXCN02000433 GALX01002836 JAB65630.1 CH964239 EDW82081.2 GANO01000692 JAB59179.1 JXUM01079660 KXJ74404.1 KA646644 AFP61273.1 UFQS01001030 UFQT01001030 SSX08629.1 SSX28545.1 CH477627 EAT38116.1 ADMH02000310 ETN67025.1 AXCM01013925 EAT36584.1 KFB41160.1 GAKP01004725 JAC54227.1 AF169229 AAD49730.1 GAKP01004726 JAC54226.1 DS233012 EDS27504.1 GGFK01011279 MBW44600.1 GDHF01004097 JAI48217.1 GDHF01011563 JAI40751.1 JXUM01079656 KXJ74402.1 ETN67026.1 CH940655 EDW66277.1 GAMC01013841 JAB92714.1 GFDF01005651 JAV08433.1 GFDF01005652 JAV08432.1 KRF99569.1 CH933812 EDW05845.1 CH379064 KRT06403.1 GBXI01017413 JAC96878.1 CH954180 EDV46098.2 AXCN02000266 GFDF01005657 JAV08427.1 JF308210 ADX78255.1 BT128502 AEE63459.1 MG770323 AXY94925.1 ADMH02002183 ETN57988.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053268
UP000053240
+ More
UP000002320 UP000075902 UP000075840 UP000069940 UP000249989 UP000075882 UP000027135 UP000075903 UP000007062 UP000008820 UP000075901 UP000076407 UP000075900 UP000075920 UP000075883 UP000235965 UP000095300 UP000075880 UP000030765 UP000076408 UP000095301 UP000075886 UP000075881 UP000007798 UP000000673 UP000008792 UP000075884 UP000075885 UP000009192 UP000001819 UP000008711
UP000002320 UP000075902 UP000075840 UP000069940 UP000249989 UP000075882 UP000027135 UP000075903 UP000007062 UP000008820 UP000075901 UP000076407 UP000075900 UP000075920 UP000075883 UP000235965 UP000095300 UP000075880 UP000030765 UP000076408 UP000095301 UP000075886 UP000075881 UP000007798 UP000000673 UP000008792 UP000075884 UP000075885 UP000009192 UP000001819 UP000008711
Pfam
Interpro
IPR029052
Metallo-depent_PP-like
+ More
IPR006179 5_nucleotidase/apyrase
IPR006146 5'-Nucleotdase_CS
IPR036907 5'-Nucleotdase_C_sf
IPR004843 Calcineurin-like_PHP_ApaH
IPR008334 5'-Nucleotdase_C
IPR005045 CDC50/LEM3_fam
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013783 Ig-like_fold
IPR013098 Ig_I-set
IPR013106 Ig_V-set
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR033116 TRYPSIN_SER
IPR009003 Peptidase_S1_PA
IPR001254 Trypsin_dom
IPR036869 J_dom_sf
IPR001623 DnaJ_domain
IPR018114 TRYPSIN_HIS
IPR007829 TM2
IPR006179 5_nucleotidase/apyrase
IPR006146 5'-Nucleotdase_CS
IPR036907 5'-Nucleotdase_C_sf
IPR004843 Calcineurin-like_PHP_ApaH
IPR008334 5'-Nucleotdase_C
IPR005045 CDC50/LEM3_fam
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013783 Ig-like_fold
IPR013098 Ig_I-set
IPR013106 Ig_V-set
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR033116 TRYPSIN_SER
IPR009003 Peptidase_S1_PA
IPR001254 Trypsin_dom
IPR036869 J_dom_sf
IPR001623 DnaJ_domain
IPR018114 TRYPSIN_HIS
IPR007829 TM2
Gene 3D
ProteinModelPortal
H9JTK9
A0A2A4JWR7
A0A2A4JW12
A0A437B966
A0A212FMH9
A0A194Q670
+ More
A0A2A4JX59 A0A194R1G7 A0A2A4JW25 A0A3S2M1V1 A0A2A4JVW4 A0A194R505 H9JL90 A0A2H1WZX1 A0A194R6M3 B0WJ00 A0A194PLJ4 A0A194PMT9 A0A1Q3FIZ4 A0A182U110 A0A2C9GRQ1 A0A182GQ46 A0A1Q3FN26 A0A1C7CZG9 A0A067RQX4 A0A182VC59 A0A1S4KDB8 Q7QIZ1 Q16QA5 A0A182HLB4 Q8T5H5 Q17NS8 A0A182L9P8 A0A182SCB1 Q8MU74 A0A182WSG4 A0A182V0W0 A0A182R636 A0A182VZA6 A0A182MGY7 A0A2J7RI11 A0A182TN56 A0A336LSN5 A0A1L8ECN7 A0A1I8PB47 A0A336LWJ0 A0A1Y1N7H1 A0A182IVB6 A0A084VT67 A0A182YKS7 A0A1I8NGR4 A0A182R0R0 A0A182K4G0 A0A182YAL5 V5G6C5 B4NEB2 U5EW72 A0A182GQ47 T1PD85 A0A336KUY8 Q16UA0 W5JVI8 A0A182M997 Q16QA4 A0A084VT66 A0A034WIT9 Q9U9I6 A0A034WFB4 B0XGC2 A0A2M4AV02 A0A0K8WAX8 A0A0K8VPE5 A0A182GQ45 W5JSD1 B4MAL8 W8BUB4 A0A182NS57 A0A1L8DPP0 A0A1L8DPP4 A0A182J0X1 A0A182JMY7 A0A182PGQ3 A0A0Q9X374 B4L649 A0A0R3P4U5 A0A182WX10 A0A0A1WD31 A0A1I8NCS8 B3NVB0 A0A182Q352 A0A1L8DPX0 F1JZ10 B3A0N5 A0A182QET4 A0A182PGQ4 A0A182PCT2 J3JZ21 A0A3G1T1Q5 W5J3U7
A0A2A4JX59 A0A194R1G7 A0A2A4JW25 A0A3S2M1V1 A0A2A4JVW4 A0A194R505 H9JL90 A0A2H1WZX1 A0A194R6M3 B0WJ00 A0A194PLJ4 A0A194PMT9 A0A1Q3FIZ4 A0A182U110 A0A2C9GRQ1 A0A182GQ46 A0A1Q3FN26 A0A1C7CZG9 A0A067RQX4 A0A182VC59 A0A1S4KDB8 Q7QIZ1 Q16QA5 A0A182HLB4 Q8T5H5 Q17NS8 A0A182L9P8 A0A182SCB1 Q8MU74 A0A182WSG4 A0A182V0W0 A0A182R636 A0A182VZA6 A0A182MGY7 A0A2J7RI11 A0A182TN56 A0A336LSN5 A0A1L8ECN7 A0A1I8PB47 A0A336LWJ0 A0A1Y1N7H1 A0A182IVB6 A0A084VT67 A0A182YKS7 A0A1I8NGR4 A0A182R0R0 A0A182K4G0 A0A182YAL5 V5G6C5 B4NEB2 U5EW72 A0A182GQ47 T1PD85 A0A336KUY8 Q16UA0 W5JVI8 A0A182M997 Q16QA4 A0A084VT66 A0A034WIT9 Q9U9I6 A0A034WFB4 B0XGC2 A0A2M4AV02 A0A0K8WAX8 A0A0K8VPE5 A0A182GQ45 W5JSD1 B4MAL8 W8BUB4 A0A182NS57 A0A1L8DPP0 A0A1L8DPP4 A0A182J0X1 A0A182JMY7 A0A182PGQ3 A0A0Q9X374 B4L649 A0A0R3P4U5 A0A182WX10 A0A0A1WD31 A0A1I8NCS8 B3NVB0 A0A182Q352 A0A1L8DPX0 F1JZ10 B3A0N5 A0A182QET4 A0A182PGQ4 A0A182PCT2 J3JZ21 A0A3G1T1Q5 W5J3U7
PDB
4H2I
E-value=4.38944e-89,
Score=838
Ontologies
GO
PANTHER
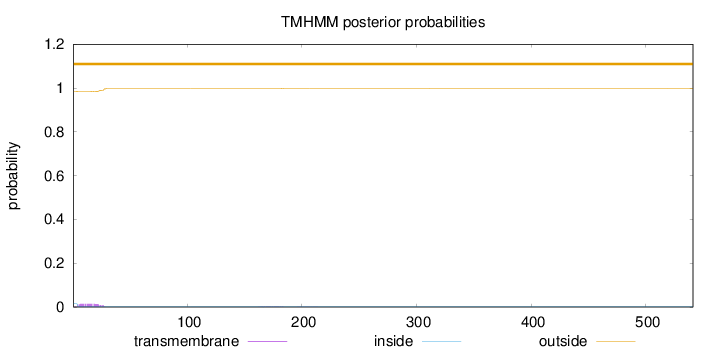
Topology
Subcellular location
Secreted
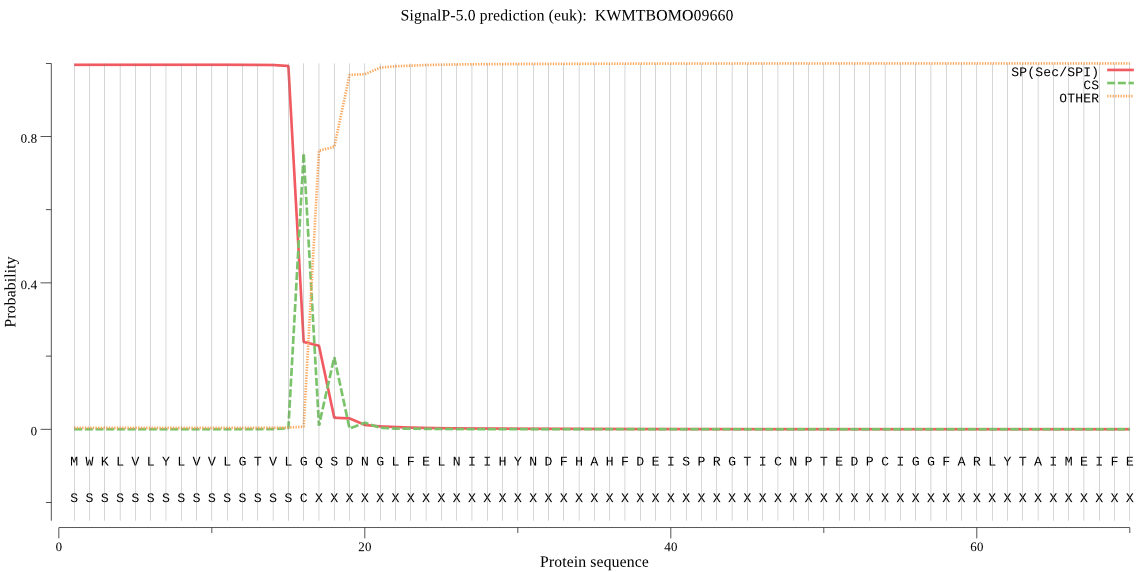
SignalP
Position: 1 - 16,
Likelihood: 0.995809
Length:
541
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.346300000000001
Exp number, first 60 AAs:
0.30384
Total prob of N-in:
0.01602
outside
1 - 541
Population Genetic Test Statistics
Pi
202.585045
Theta
150.376191
Tajima's D
1.032896
CLR
0.242947
CSRT
0.668066596670167
Interpretation
Uncertain
