Gene
KWMTBOMO09659
Pre Gene Modal
BGIBMGA012870
Annotation
hypothetical_protein_KGM_15486_[Danaus_plexippus]
Location in the cell
PlasmaMembrane Reliability : 2.588
Sequence
CDS
ATGGCGACGTCTAGTGATACGTCGGAGCAAAATGTAAAGTCAAGACGACCTGCTGAATCTGCTTTTAAGCAGCAACGTCTACCTGCATGGCAACCAATACTGACTGCTGGAACTGTGCTACCTACGTTCTTTGTAATCGGAATCGCTTTTATACCAGTCGGGATAGGTCTGCTCTATTTTTCTGATGAGGTTAAAGAGCATGTTATTGACTACACTTATTGCATGAAAGATGATATGAACGTCACATGTGCAGATTTTTTAAAAAACAACACTGAAGAAATATGCACTTGTCATCTTCCCTTCAATCTTACTGAGGACTTCAAAGGAGAAGTTTATTTCTATTATGGACTGACCAATTATTACCAGAATCATAGACGATATGTAAAATCCAGGGATGACAATCAGCTTTTAGGCCGCCTGTCATTAACTCCATCTTCTGATTGTGATCCATTTGCACGTGCTGAAGAAAATGGAGTAATGAAACCAATAGCACCTTGTGGAGCTATAGCTAACTCTTTGTTCAATGATACACTTACATTGCATTCAGTGGAATCGAATGGCGATGTTCCTGTACTACGAACTGGTATTGCCTGGGCATCTGATAAGAGGACCAAGTTCCGGAACCCAGAAGGCAATCTCGAAGAAGCATTTGCAAACTTTACGAAGCCAGTGAATTGGCGTGTCAACATTTGGCAGTTAGATCCCGGCAATCCTGATAACAACGGTTTTCAGAATGAAGACTTGATCGTGTGGATGCGCACAGCCGCCCTGCCGACATTCCGCAAGTTGTATCGGCGCGTGGACCACAGCCAGGTAGGGTTCGGTGGTGGACTCGTCAAAGGCCCGTATATTCTCAAAGTCGACTACAATTACCCAGTAGTTGACTTCCATGGGACGAAATCATTTATAATTTCGACTACATCGTTGCTGGGGGGAAAGAATCCGTTCTTAGGAGTGGCTTATGTAGTCGTCGGTACCCTGTGCCTCTTACTGGGCATAGTACTACTCGTCATACACGTCAGATGTTCCAAGAGCACAACCGAAATGATCAACGTGAACCCGCGCACGCCGTACTCTTGA
Protein
MATSSDTSEQNVKSRRPAESAFKQQRLPAWQPILTAGTVLPTFFVIGIAFIPVGIGLLYFSDEVKEHVIDYTYCMKDDMNVTCADFLKNNTEEICTCHLPFNLTEDFKGEVYFYYGLTNYYQNHRRYVKSRDDNQLLGRLSLTPSSDCDPFARAEENGVMKPIAPCGAIANSLFNDTLTLHSVESNGDVPVLRTGIAWASDKRTKFRNPEGNLEEAFANFTKPVNWRVNIWQLDPGNPDNNGFQNEDLIVWMRTAALPTFRKLYRRVDHSQVGFGGGLVKGPYILKVDYNYPVVDFHGTKSFIISTTSLLGGKNPFLGVAYVVVGTLCLLLGIVLLVIHVRCSKSTTEMINVNPRTPYS
Summary
Similarity
Belongs to the CDC50/LEM3 family.
Uniprot
A0A2A4JXJ5
A0A212FMG5
A0A3S2LHQ0
A0A194Q670
A0A2H1VHJ0
A0A1E1W467
+ More
H9JTK8 W8C150 A0A0K8UI11 A0A067QYJ3 A0A0A1WY30 A0A034WFH4 A0A1L8DWD3 A0A158NTV6 V5G7H2 A0A1I8M348 F4WJ03 A0A1B6EAH3 A0A1L8EE67 A0A1L8EE62 A0A0K8TP30 D6WZE8 A0A1A9VWY8 A0A1I8PI20 A0A1A9YGT2 A0A1B0BGL0 K7IN94 A0A1A9WFQ6 B4L5N6 A0A087ZQI6 D3TS55 A0A0C9RDN0 A0A3L8DDE8 B4M341 A0A0L0BRN1 A0A026WP77 A0A195BP23 A0A2A3E421 A0A0A9WLT9 A0A0M4EW75 A0A336N1R8 A0A2J7QRW3 A0A2J7QRV7 U5ETG0 A0A1Q3FF83 B0XHV9 Q9VXG0 E2BFV4 Q8STD2 A0A3B0JQC6 A0A1W4VJI3 Q17J53 A0A0P4VLT1 R4FNF2 A0A3B0JY48 B3NX37 A0A0T6B1P2 A0A1Y1NM88 A0A151J3X1 B4PXT5 A0A1W4VWS8 A0A0J7KQW2 B4N239 B4GXG2 V9IJG2 A0A195CTM3 A0A1L8EAS6 A0A182G523 A0A023ERP2 N6UF05 A0A023EPE8 A0A182GUJ6 A0A182FM91 E2AIR1 Q29HM2 A0A0P5T661 A0A0P5ZHU0 A0A0P6EJN0 A0A0P5PKH2 B3MXT7 A0A069DT93 A0A0L7QXV4 A0A1B6K552 A0A0R3NZP8 A0A0P6IE38 X2JKP3 J3JVE0 A0A3B0JC59 A0A1B6GFZ0 A0A0P5BW51 A0A2M4BSX2 W5JH39 A0A2R7W982 A0A0P4Y1W0 A0A224XF55 A0A2M3Z277 A0A2M4ADG9 E9H1A0 A0A0V0G3H7 A0A2P8Y9U9
H9JTK8 W8C150 A0A0K8UI11 A0A067QYJ3 A0A0A1WY30 A0A034WFH4 A0A1L8DWD3 A0A158NTV6 V5G7H2 A0A1I8M348 F4WJ03 A0A1B6EAH3 A0A1L8EE67 A0A1L8EE62 A0A0K8TP30 D6WZE8 A0A1A9VWY8 A0A1I8PI20 A0A1A9YGT2 A0A1B0BGL0 K7IN94 A0A1A9WFQ6 B4L5N6 A0A087ZQI6 D3TS55 A0A0C9RDN0 A0A3L8DDE8 B4M341 A0A0L0BRN1 A0A026WP77 A0A195BP23 A0A2A3E421 A0A0A9WLT9 A0A0M4EW75 A0A336N1R8 A0A2J7QRW3 A0A2J7QRV7 U5ETG0 A0A1Q3FF83 B0XHV9 Q9VXG0 E2BFV4 Q8STD2 A0A3B0JQC6 A0A1W4VJI3 Q17J53 A0A0P4VLT1 R4FNF2 A0A3B0JY48 B3NX37 A0A0T6B1P2 A0A1Y1NM88 A0A151J3X1 B4PXT5 A0A1W4VWS8 A0A0J7KQW2 B4N239 B4GXG2 V9IJG2 A0A195CTM3 A0A1L8EAS6 A0A182G523 A0A023ERP2 N6UF05 A0A023EPE8 A0A182GUJ6 A0A182FM91 E2AIR1 Q29HM2 A0A0P5T661 A0A0P5ZHU0 A0A0P6EJN0 A0A0P5PKH2 B3MXT7 A0A069DT93 A0A0L7QXV4 A0A1B6K552 A0A0R3NZP8 A0A0P6IE38 X2JKP3 J3JVE0 A0A3B0JC59 A0A1B6GFZ0 A0A0P5BW51 A0A2M4BSX2 W5JH39 A0A2R7W982 A0A0P4Y1W0 A0A224XF55 A0A2M3Z277 A0A2M4ADG9 E9H1A0 A0A0V0G3H7 A0A2P8Y9U9
Pubmed
22118469
26354079
19121390
24495485
24845553
25830018
+ More
25348373 21347285 25315136 21719571 26369729 18362917 19820115 20075255 17994087 20353571 30249741 26108605 24508170 25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 17510324 27129103 28004739 17550304 26483478 24945155 23537049 15632085 26334808 22516182 20920257 23761445 21292972 29403074
25348373 21347285 25315136 21719571 26369729 18362917 19820115 20075255 17994087 20353571 30249741 26108605 24508170 25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 17510324 27129103 28004739 17550304 26483478 24945155 23537049 15632085 26334808 22516182 20920257 23761445 21292972 29403074
EMBL
NWSH01000463
PCG76223.1
AGBW02007651
OWR54938.1
RSAL01000115
RVE46872.1
+ More
KQ459582 KPI98905.1 ODYU01002588 SOQ40285.1 GDQN01009276 JAT81778.1 BABH01003659 GAMC01011141 JAB95414.1 GDHF01026118 JAI26196.1 KK852822 KDR15530.1 GBXI01010545 JAD03747.1 GAKP01006424 JAC52528.1 GFDF01003338 JAV10746.1 ADTU01026111 GALX01002386 JAB66080.1 GL888181 EGI65773.1 GEDC01002369 JAS34929.1 GFDG01001808 JAV16991.1 GFDG01001811 JAV16988.1 GDAI01001680 JAI15923.1 KQ971372 EFA10416.1 JXJN01013888 CH933811 EDW06495.2 CCAG010001343 EZ424257 ADD20533.1 GBYB01003567 GBYB01011212 GBYB01011265 JAG73334.1 JAG80979.1 JAG81032.1 QOIP01000009 RLU18414.1 CH940651 EDW65216.2 JRES01001467 KNC22681.1 KK107148 EZA57466.1 KQ976435 KYM87107.1 KZ288386 PBC26477.1 GBHO01044711 GBHO01039746 GBHO01034880 GBRD01017000 GBRD01016026 GBRD01016025 GBRD01016024 GBRD01012206 GBRD01005452 GBRD01000837 JAF98892.1 JAG03858.1 JAG08724.1 JAG48827.1 CP012528 ALC49660.1 UFQT01003361 SSX34903.1 NEVH01011884 PNF31320.1 PNF31318.1 GANO01002800 JAB57071.1 GFDL01008835 JAV26210.1 DS233218 EDS28768.1 AE014298 BT044187 AAF48613.1 ACH92252.1 GL448096 EFN85356.1 AY070651 AY070652 AAL48122.1 AAL48123.1 OUUW01000003 SPP77650.1 CH477235 EAT46656.1 GDKW01001191 JAI55404.1 ACPB03010896 GAHY01001319 JAA76191.1 SPP77651.1 CH954180 EDV46866.1 LJIG01016216 KRT81266.1 GEZM01002070 JAV97386.1 KQ980246 KYN17128.1 CM000162 EDX01921.1 LBMM01004223 KMQ92681.1 CH963925 EDW78428.2 CH479196 EDW27439.1 JR048225 AEY60606.1 KQ977279 KYN04041.1 GFDG01002978 JAV15821.1 JXUM01042851 JXUM01042852 JXUM01042853 JXUM01042854 KQ561341 KXJ78912.1 GAPW01002242 JAC11356.1 APGK01029568 KB740735 KB632302 ENN79206.1 ERL91778.1 GAPW01002241 JAC11357.1 JXUM01018320 JXUM01018321 JXUM01018322 JXUM01018323 JXUM01018324 JXUM01018325 JXUM01018326 KQ560509 KXJ82069.1 GL439869 EFN66673.1 CH379064 EAL31736.1 GDIP01130456 JAL73258.1 GDIP01043772 JAM59943.1 GDIQ01068671 LRGB01000024 JAN26066.1 KZS21685.1 GDIQ01127150 JAL24576.1 CH902630 EDV38552.2 GBGD01002032 JAC86857.1 KQ414699 KOC63447.1 GECU01001138 JAT06569.1 KRT06498.1 GDIQ01005880 JAN88857.1 AHN59797.1 BT127207 AEE62169.1 SPP77652.1 GECZ01008422 JAS61347.1 GDIP01183644 JAJ39758.1 GGFJ01006727 MBW55868.1 ADMH02001411 ETN62638.1 KK854481 PTY16243.1 GDIP01234992 JAI88409.1 GFTR01005309 JAW11117.1 GGFM01001869 MBW22620.1 GGFK01005502 MBW38823.1 GL732583 EFX74453.1 GECL01003507 JAP02617.1 PYGN01000771 PSN41029.1
KQ459582 KPI98905.1 ODYU01002588 SOQ40285.1 GDQN01009276 JAT81778.1 BABH01003659 GAMC01011141 JAB95414.1 GDHF01026118 JAI26196.1 KK852822 KDR15530.1 GBXI01010545 JAD03747.1 GAKP01006424 JAC52528.1 GFDF01003338 JAV10746.1 ADTU01026111 GALX01002386 JAB66080.1 GL888181 EGI65773.1 GEDC01002369 JAS34929.1 GFDG01001808 JAV16991.1 GFDG01001811 JAV16988.1 GDAI01001680 JAI15923.1 KQ971372 EFA10416.1 JXJN01013888 CH933811 EDW06495.2 CCAG010001343 EZ424257 ADD20533.1 GBYB01003567 GBYB01011212 GBYB01011265 JAG73334.1 JAG80979.1 JAG81032.1 QOIP01000009 RLU18414.1 CH940651 EDW65216.2 JRES01001467 KNC22681.1 KK107148 EZA57466.1 KQ976435 KYM87107.1 KZ288386 PBC26477.1 GBHO01044711 GBHO01039746 GBHO01034880 GBRD01017000 GBRD01016026 GBRD01016025 GBRD01016024 GBRD01012206 GBRD01005452 GBRD01000837 JAF98892.1 JAG03858.1 JAG08724.1 JAG48827.1 CP012528 ALC49660.1 UFQT01003361 SSX34903.1 NEVH01011884 PNF31320.1 PNF31318.1 GANO01002800 JAB57071.1 GFDL01008835 JAV26210.1 DS233218 EDS28768.1 AE014298 BT044187 AAF48613.1 ACH92252.1 GL448096 EFN85356.1 AY070651 AY070652 AAL48122.1 AAL48123.1 OUUW01000003 SPP77650.1 CH477235 EAT46656.1 GDKW01001191 JAI55404.1 ACPB03010896 GAHY01001319 JAA76191.1 SPP77651.1 CH954180 EDV46866.1 LJIG01016216 KRT81266.1 GEZM01002070 JAV97386.1 KQ980246 KYN17128.1 CM000162 EDX01921.1 LBMM01004223 KMQ92681.1 CH963925 EDW78428.2 CH479196 EDW27439.1 JR048225 AEY60606.1 KQ977279 KYN04041.1 GFDG01002978 JAV15821.1 JXUM01042851 JXUM01042852 JXUM01042853 JXUM01042854 KQ561341 KXJ78912.1 GAPW01002242 JAC11356.1 APGK01029568 KB740735 KB632302 ENN79206.1 ERL91778.1 GAPW01002241 JAC11357.1 JXUM01018320 JXUM01018321 JXUM01018322 JXUM01018323 JXUM01018324 JXUM01018325 JXUM01018326 KQ560509 KXJ82069.1 GL439869 EFN66673.1 CH379064 EAL31736.1 GDIP01130456 JAL73258.1 GDIP01043772 JAM59943.1 GDIQ01068671 LRGB01000024 JAN26066.1 KZS21685.1 GDIQ01127150 JAL24576.1 CH902630 EDV38552.2 GBGD01002032 JAC86857.1 KQ414699 KOC63447.1 GECU01001138 JAT06569.1 KRT06498.1 GDIQ01005880 JAN88857.1 AHN59797.1 BT127207 AEE62169.1 SPP77652.1 GECZ01008422 JAS61347.1 GDIP01183644 JAJ39758.1 GGFJ01006727 MBW55868.1 ADMH02001411 ETN62638.1 KK854481 PTY16243.1 GDIP01234992 JAI88409.1 GFTR01005309 JAW11117.1 GGFM01001869 MBW22620.1 GGFK01005502 MBW38823.1 GL732583 EFX74453.1 GECL01003507 JAP02617.1 PYGN01000771 PSN41029.1
Proteomes
UP000218220
UP000007151
UP000283053
UP000053268
UP000005204
UP000027135
+ More
UP000005205 UP000095301 UP000007755 UP000007266 UP000078200 UP000095300 UP000092443 UP000092460 UP000002358 UP000091820 UP000009192 UP000005203 UP000092444 UP000279307 UP000008792 UP000037069 UP000053097 UP000078540 UP000242457 UP000092553 UP000235965 UP000002320 UP000000803 UP000008237 UP000268350 UP000192221 UP000008820 UP000015103 UP000008711 UP000078492 UP000002282 UP000036403 UP000007798 UP000008744 UP000078542 UP000069940 UP000249989 UP000019118 UP000030742 UP000069272 UP000000311 UP000001819 UP000076858 UP000007801 UP000053825 UP000000673 UP000000305 UP000245037
UP000005205 UP000095301 UP000007755 UP000007266 UP000078200 UP000095300 UP000092443 UP000092460 UP000002358 UP000091820 UP000009192 UP000005203 UP000092444 UP000279307 UP000008792 UP000037069 UP000053097 UP000078540 UP000242457 UP000092553 UP000235965 UP000002320 UP000000803 UP000008237 UP000268350 UP000192221 UP000008820 UP000015103 UP000008711 UP000078492 UP000002282 UP000036403 UP000007798 UP000008744 UP000078542 UP000069940 UP000249989 UP000019118 UP000030742 UP000069272 UP000000311 UP000001819 UP000076858 UP000007801 UP000053825 UP000000673 UP000000305 UP000245037
Interpro
SUPFAM
SSF55816
SSF55816
Gene 3D
ProteinModelPortal
A0A2A4JXJ5
A0A212FMG5
A0A3S2LHQ0
A0A194Q670
A0A2H1VHJ0
A0A1E1W467
+ More
H9JTK8 W8C150 A0A0K8UI11 A0A067QYJ3 A0A0A1WY30 A0A034WFH4 A0A1L8DWD3 A0A158NTV6 V5G7H2 A0A1I8M348 F4WJ03 A0A1B6EAH3 A0A1L8EE67 A0A1L8EE62 A0A0K8TP30 D6WZE8 A0A1A9VWY8 A0A1I8PI20 A0A1A9YGT2 A0A1B0BGL0 K7IN94 A0A1A9WFQ6 B4L5N6 A0A087ZQI6 D3TS55 A0A0C9RDN0 A0A3L8DDE8 B4M341 A0A0L0BRN1 A0A026WP77 A0A195BP23 A0A2A3E421 A0A0A9WLT9 A0A0M4EW75 A0A336N1R8 A0A2J7QRW3 A0A2J7QRV7 U5ETG0 A0A1Q3FF83 B0XHV9 Q9VXG0 E2BFV4 Q8STD2 A0A3B0JQC6 A0A1W4VJI3 Q17J53 A0A0P4VLT1 R4FNF2 A0A3B0JY48 B3NX37 A0A0T6B1P2 A0A1Y1NM88 A0A151J3X1 B4PXT5 A0A1W4VWS8 A0A0J7KQW2 B4N239 B4GXG2 V9IJG2 A0A195CTM3 A0A1L8EAS6 A0A182G523 A0A023ERP2 N6UF05 A0A023EPE8 A0A182GUJ6 A0A182FM91 E2AIR1 Q29HM2 A0A0P5T661 A0A0P5ZHU0 A0A0P6EJN0 A0A0P5PKH2 B3MXT7 A0A069DT93 A0A0L7QXV4 A0A1B6K552 A0A0R3NZP8 A0A0P6IE38 X2JKP3 J3JVE0 A0A3B0JC59 A0A1B6GFZ0 A0A0P5BW51 A0A2M4BSX2 W5JH39 A0A2R7W982 A0A0P4Y1W0 A0A224XF55 A0A2M3Z277 A0A2M4ADG9 E9H1A0 A0A0V0G3H7 A0A2P8Y9U9
H9JTK8 W8C150 A0A0K8UI11 A0A067QYJ3 A0A0A1WY30 A0A034WFH4 A0A1L8DWD3 A0A158NTV6 V5G7H2 A0A1I8M348 F4WJ03 A0A1B6EAH3 A0A1L8EE67 A0A1L8EE62 A0A0K8TP30 D6WZE8 A0A1A9VWY8 A0A1I8PI20 A0A1A9YGT2 A0A1B0BGL0 K7IN94 A0A1A9WFQ6 B4L5N6 A0A087ZQI6 D3TS55 A0A0C9RDN0 A0A3L8DDE8 B4M341 A0A0L0BRN1 A0A026WP77 A0A195BP23 A0A2A3E421 A0A0A9WLT9 A0A0M4EW75 A0A336N1R8 A0A2J7QRW3 A0A2J7QRV7 U5ETG0 A0A1Q3FF83 B0XHV9 Q9VXG0 E2BFV4 Q8STD2 A0A3B0JQC6 A0A1W4VJI3 Q17J53 A0A0P4VLT1 R4FNF2 A0A3B0JY48 B3NX37 A0A0T6B1P2 A0A1Y1NM88 A0A151J3X1 B4PXT5 A0A1W4VWS8 A0A0J7KQW2 B4N239 B4GXG2 V9IJG2 A0A195CTM3 A0A1L8EAS6 A0A182G523 A0A023ERP2 N6UF05 A0A023EPE8 A0A182GUJ6 A0A182FM91 E2AIR1 Q29HM2 A0A0P5T661 A0A0P5ZHU0 A0A0P6EJN0 A0A0P5PKH2 B3MXT7 A0A069DT93 A0A0L7QXV4 A0A1B6K552 A0A0R3NZP8 A0A0P6IE38 X2JKP3 J3JVE0 A0A3B0JC59 A0A1B6GFZ0 A0A0P5BW51 A0A2M4BSX2 W5JH39 A0A2R7W982 A0A0P4Y1W0 A0A224XF55 A0A2M3Z277 A0A2M4ADG9 E9H1A0 A0A0V0G3H7 A0A2P8Y9U9
Ontologies
GO
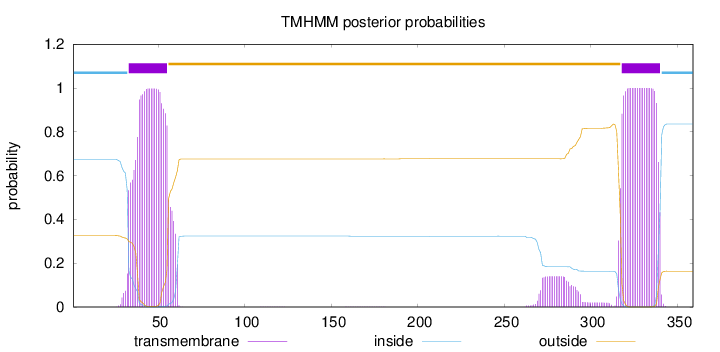
Topology
Length:
359
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
48.94016
Exp number, first 60 AAs:
22.58771
Total prob of N-in:
0.67378
POSSIBLE N-term signal
sequence
inside
1 - 32
TMhelix
33 - 55
outside
56 - 317
TMhelix
318 - 340
inside
341 - 359
Population Genetic Test Statistics
Pi
204.294503
Theta
151.786811
Tajima's D
0.515566
CLR
0.005399
CSRT
0.517874106294685
Interpretation
Uncertain
