Pre Gene Modal
BGIBMGA012966
Annotation
septin_[Bombyx_mori]
Full name
Septin-1
Alternative Name
DIFF6 protein homolog
Protein innocent bystander
Protein innocent bystander
Location in the cell
Cytoplasmic Reliability : 2.177 Nuclear Reliability : 2.282
Sequence
CDS
ATGTCAAATGAAACAAATAAAACATTCTCAAATCTGGAAACACCTGGTTATGTTGGATTTGCCAATCTGCCTAATCAGGTACACAGAAAATCTGTAAAAAAGGGTTTCGAGTTCACCCTTATGGTGGTCGGGGAAAGTGGACTGGGCAAATCTACTCTGGTCAACTCTTTGTTCTTGACCGATTTATACCCTGAACGAGTAATCCCTGATGCTACTGAGAAAACAAACCAGACCGTGAAATTAGACGCATCGACGGTCGAGATAGAAGAGCGCGGCGTGAAGCTGCGTTTGACCGTCGTGGACACGCCCGGCTACGGAGACGCGATTGATAACACGGATTGTTTTCGATCCATAATTCAATACATTGACGAGCAGTTCGAAAGATTTCTTCGAGACGAGAGCGGCCTAAACCGTCGTAACATTGTGGACAATCGCATTCATTGCTGTTTCTACTTCATATCACCGTTTGGACACGGGTTAAAGCCTCTTGATATTGAGTTCATGAAACAGCTGCACAACAAAGTTAACATCGTACCGGTTATCGCCAAAGCGGACTGTCTCACTAAGAAAGAAGTTCAGCGACTGAAATCTAGGGTTATGGAAGAAATCGAAAGGGAAGGCATAAAGATATACCCGCTGCCCGATTGCGACAGTGACGAAGACGAAGATTACAAAGAACAGGTCCGCCAGCTGAAGGAGGCGGTGCCGTTCGCAGTGTGCGGCGCGGGCCAGCAGCTGGAGGTGCGCGGGCGGCGGGTGCGCGGCCGCCTCTACCCCTGGGGAGTCGTCGAGGTCGAGAACCCTGAGCACTGCGACTTCATCAAACTCAGGACCATGCTCATTACCCACATGCAGGACCTCCAAGAGGTCACACAGGAAGTTCACTATGAGAACTATCGTTCGGAGAGACTCGCCCGCACCGGCCAAGTTCCGAAACGCCACACGTCAACTGAAAGCGGACTTAGTGAGGCGGACTCTGCGCTCACCAATGGCTCCGCGGAAGACTCCACCGAGCGCGAGCGTGCCCTACGTGAAAAGGAGGCAGAATTACGTCGCATGCAAGAGATGTTGGAGCAGATGCAGCGGCAAATGCAATTGCAGGCCGCCGGTGCTACCACCGCCACCTCAACCACTGCATAG
Protein
MSNETNKTFSNLETPGYVGFANLPNQVHRKSVKKGFEFTLMVVGESGLGKSTLVNSLFLTDLYPERVIPDATEKTNQTVKLDASTVEIEERGVKLRLTVVDTPGYGDAIDNTDCFRSIIQYIDEQFERFLRDESGLNRRNIVDNRIHCCFYFISPFGHGLKPLDIEFMKQLHNKVNIVPVIAKADCLTKKEVQRLKSRVMEEIEREGIKIYPLPDCDSDEDEDYKEQVRQLKEAVPFAVCGAGQQLEVRGRRVRGRLYPWGVVEVENPEHCDFIKLRTMLITHMQDLQEVTQEVHYENYRSERLARTGQVPKRHTSTESGLSEADSALTNGSAEDSTERERALREKEAELRRMQEMLEQMQRQMQLQAAGATTATSTTA
Summary
Description
Involved in cytokinesis.
Subunit
May assemble into a multicomponent structure.
Similarity
Belongs to the TRAFAC class TrmE-Era-EngA-EngB-Septin-like GTPase superfamily. Septin GTPase family.
Keywords
Cell cycle
Cell division
Complete proteome
GTP-binding
Nucleotide-binding
Phosphoprotein
Reference proteome
Feature
chain Septin-1
Uniprot
Q1HQB2
A0A212FMM4
A0A194R2R4
A0A194Q015
A0A437B8K6
A0A2A4JXT8
+ More
H9JTV4 F4WRF2 A0A195BE60 A0A158NUL1 A0A2A3EM85 A0A088AKH0 A0A2J7PGQ1 A0A067RHK0 K7J9N2 A0A026W8T3 A0A1S4N210 E2AFV7 E0W3N0 A0A151JQY4 A0A154PRU7 A0A195FJS2 A0A1B6DLZ5 A0A151IFQ3 A0A1W4WZ12 A0A1B6F7U1 A0A0N0U609 A0A0C9Q809 A0A3L8DL59 A0A2S2Q0M9 A0A2H8TPM8 A0A2S2PUK1 A0A1Y1KLI9 W8AZP6 J9K1P5 A0A182H1D8 A0A0A9YUY9 A0A1Q3FWK3 A0A0P4VMD8 A0A0R3NXT6 A0A0K8VHJ0 A0A034VBT2 A0A224XF00 D2A6H6 A0A069DYQ8 A0A0K8S8Y1 A0A0A1WR70 Q29JE4 B4H9X4 A0A0R3NXX1 A0A3B0JQQ1 R4FR01 A0A023F9C1 T1P9J6 B4L5D8 A0A1L8DXH0 A0A1I8Q8V8 A0A1W4UVS1 B4I6H1 A0A1A9W191 B4MAZ6 B3MRW1 A0A182XAK2 A0A0L0BVQ7 A0A182HSE2 Q7QCI4 A0A1I8MCR5 A0A1A9XC23 A0A1A9VB08 A0A0Q9WMC8 A0A1I8Q8T0 A0A2M3ZJH8 A0A1B0A9Y6 A0A2M4ATS3 A0A2M4BSG1 A0A182QGP8 A0A0M5JAL8 A0A182Y6N6 B4JNF9 A0A182FAJ0 W5JK94 A0A1S4FB03 A0A0T6B0R0 A0A182P3T8 Q179W2 A0A182NRB5 Q540V5 U5EUH0 B4PZ33 B3NYD7 P42207 A0A1B0BGK8 A0A182IZ29 A0A084W756 A0A182RQJ2 A0A182W6A5 A0A182W2I2 A0A182LED8 A0A182JP60 A0A182TXN4
H9JTV4 F4WRF2 A0A195BE60 A0A158NUL1 A0A2A3EM85 A0A088AKH0 A0A2J7PGQ1 A0A067RHK0 K7J9N2 A0A026W8T3 A0A1S4N210 E2AFV7 E0W3N0 A0A151JQY4 A0A154PRU7 A0A195FJS2 A0A1B6DLZ5 A0A151IFQ3 A0A1W4WZ12 A0A1B6F7U1 A0A0N0U609 A0A0C9Q809 A0A3L8DL59 A0A2S2Q0M9 A0A2H8TPM8 A0A2S2PUK1 A0A1Y1KLI9 W8AZP6 J9K1P5 A0A182H1D8 A0A0A9YUY9 A0A1Q3FWK3 A0A0P4VMD8 A0A0R3NXT6 A0A0K8VHJ0 A0A034VBT2 A0A224XF00 D2A6H6 A0A069DYQ8 A0A0K8S8Y1 A0A0A1WR70 Q29JE4 B4H9X4 A0A0R3NXX1 A0A3B0JQQ1 R4FR01 A0A023F9C1 T1P9J6 B4L5D8 A0A1L8DXH0 A0A1I8Q8V8 A0A1W4UVS1 B4I6H1 A0A1A9W191 B4MAZ6 B3MRW1 A0A182XAK2 A0A0L0BVQ7 A0A182HSE2 Q7QCI4 A0A1I8MCR5 A0A1A9XC23 A0A1A9VB08 A0A0Q9WMC8 A0A1I8Q8T0 A0A2M3ZJH8 A0A1B0A9Y6 A0A2M4ATS3 A0A2M4BSG1 A0A182QGP8 A0A0M5JAL8 A0A182Y6N6 B4JNF9 A0A182FAJ0 W5JK94 A0A1S4FB03 A0A0T6B0R0 A0A182P3T8 Q179W2 A0A182NRB5 Q540V5 U5EUH0 B4PZ33 B3NYD7 P42207 A0A1B0BGK8 A0A182IZ29 A0A084W756 A0A182RQJ2 A0A182W6A5 A0A182W2I2 A0A182LED8 A0A182JP60 A0A182TXN4
Pubmed
22118469
26354079
19121390
21719571
21347285
24845553
+ More
20075255 24508170 20566863 20798317 30249741 28004739 24495485 26483478 25401762 26823975 27129103 15632085 17994087 25348373 18362917 19820115 26334808 25830018 25474469 25315136 26108605 12364791 14747013 17210077 25244985 20920257 23761445 17510324 17550304 8590810 9520435 10731132 12537572 17372656 24438588 20966253
20075255 24508170 20566863 20798317 30249741 28004739 24495485 26483478 25401762 26823975 27129103 15632085 17994087 25348373 18362917 19820115 26334808 25830018 25474469 25315136 26108605 12364791 14747013 17210077 25244985 20920257 23761445 17510324 17550304 8590810 9520435 10731132 12537572 17372656 24438588 20966253
EMBL
DQ443140
ABF51229.1
AGBW02007651
OWR54939.1
KQ460878
KPJ11540.1
+ More
KQ459582 KPI98906.1 RSAL01000115 RVE46871.1 NWSH01000463 PCG76240.1 BABH01003660 GL888285 EGI63223.1 KQ976504 KYM82871.1 ADTU01026592 KZ288217 PBC32392.1 NEVH01025174 PNF15504.1 KK852680 KDR18628.1 AAZX01002711 KK107390 EZA51439.1 AAZO01007428 GL439158 EFN67706.1 DS235882 EEB20236.1 KQ978660 KYN29387.1 KQ435095 KZC14629.1 KQ981522 KYN40611.1 GEDC01028961 GEDC01010599 JAS08337.1 JAS26699.1 KQ977771 KYN00029.1 GECZ01023482 JAS46287.1 KQ435759 KOX75671.1 GBYB01010295 GBYB01010297 GBYB01010298 JAG80062.1 JAG80064.1 JAG80065.1 QOIP01000007 RLU20933.1 GGMS01002115 MBY71318.1 GFXV01004145 MBW15950.1 GGMR01020399 MBY33018.1 GEZM01080253 JAV62273.1 GAMC01016217 JAB90338.1 ABLF02016818 JXUM01003458 JXUM01003459 JXUM01100848 JXUM01100849 JXUM01100850 KQ564603 KQ560158 KXJ72037.1 KXJ84126.1 GBHO01008178 GBHO01008177 GBHO01008176 GBHO01008175 GBHO01008174 GBHO01008172 GBHO01003148 GDHC01001252 JAG35426.1 JAG35427.1 JAG35428.1 JAG35429.1 JAG35430.1 JAG35432.1 JAG40456.1 JAQ17377.1 GFDL01003044 JAV32001.1 GDKW01002982 JAI53613.1 CH379063 KRT05911.1 GDHF01013991 JAI38323.1 GAKP01019743 GAKP01019742 GAKP01019741 GAKP01019740 GAKP01019739 JAC39210.1 GFTR01005369 JAW11057.1 KQ971348 EFA04919.1 GBGD01001965 JAC86924.1 GBRD01016088 JAG49738.1 GBXI01012990 JAD01302.1 EAL32357.2 CH479232 EDW36631.1 KRT05912.1 OUUW01000003 SPP77810.1 GAHY01000139 JAA77371.1 GBBI01000615 JAC18097.1 KA645374 AFP60003.1 CH933811 EDW06397.2 GFDF01003129 JAV10955.1 CH480823 EDW56377.1 CH940655 EDW66405.2 CH902622 EDV34516.1 JRES01001368 KNC23324.1 APCN01000266 AAAB01008859 EAA08200.4 KRF82673.1 GGFM01007935 MBW28686.1 GGFK01010884 MBW44205.1 GGFJ01006885 MBW56026.1 AXCN02001467 CP012528 ALC49036.1 CH916371 EDV92252.1 ADMH02000875 ETN64797.1 LJIG01016320 KRT80956.1 CH477344 EAT43036.1 AY121679 AAM52006.1 GANO01001473 JAB58398.1 CM000162 EDX03094.1 CH954180 EDV47616.1 L33246 X67202 AF017777 AE014298 JXJN01013884 ATLV01021113 KE525312 KFB46050.1
KQ459582 KPI98906.1 RSAL01000115 RVE46871.1 NWSH01000463 PCG76240.1 BABH01003660 GL888285 EGI63223.1 KQ976504 KYM82871.1 ADTU01026592 KZ288217 PBC32392.1 NEVH01025174 PNF15504.1 KK852680 KDR18628.1 AAZX01002711 KK107390 EZA51439.1 AAZO01007428 GL439158 EFN67706.1 DS235882 EEB20236.1 KQ978660 KYN29387.1 KQ435095 KZC14629.1 KQ981522 KYN40611.1 GEDC01028961 GEDC01010599 JAS08337.1 JAS26699.1 KQ977771 KYN00029.1 GECZ01023482 JAS46287.1 KQ435759 KOX75671.1 GBYB01010295 GBYB01010297 GBYB01010298 JAG80062.1 JAG80064.1 JAG80065.1 QOIP01000007 RLU20933.1 GGMS01002115 MBY71318.1 GFXV01004145 MBW15950.1 GGMR01020399 MBY33018.1 GEZM01080253 JAV62273.1 GAMC01016217 JAB90338.1 ABLF02016818 JXUM01003458 JXUM01003459 JXUM01100848 JXUM01100849 JXUM01100850 KQ564603 KQ560158 KXJ72037.1 KXJ84126.1 GBHO01008178 GBHO01008177 GBHO01008176 GBHO01008175 GBHO01008174 GBHO01008172 GBHO01003148 GDHC01001252 JAG35426.1 JAG35427.1 JAG35428.1 JAG35429.1 JAG35430.1 JAG35432.1 JAG40456.1 JAQ17377.1 GFDL01003044 JAV32001.1 GDKW01002982 JAI53613.1 CH379063 KRT05911.1 GDHF01013991 JAI38323.1 GAKP01019743 GAKP01019742 GAKP01019741 GAKP01019740 GAKP01019739 JAC39210.1 GFTR01005369 JAW11057.1 KQ971348 EFA04919.1 GBGD01001965 JAC86924.1 GBRD01016088 JAG49738.1 GBXI01012990 JAD01302.1 EAL32357.2 CH479232 EDW36631.1 KRT05912.1 OUUW01000003 SPP77810.1 GAHY01000139 JAA77371.1 GBBI01000615 JAC18097.1 KA645374 AFP60003.1 CH933811 EDW06397.2 GFDF01003129 JAV10955.1 CH480823 EDW56377.1 CH940655 EDW66405.2 CH902622 EDV34516.1 JRES01001368 KNC23324.1 APCN01000266 AAAB01008859 EAA08200.4 KRF82673.1 GGFM01007935 MBW28686.1 GGFK01010884 MBW44205.1 GGFJ01006885 MBW56026.1 AXCN02001467 CP012528 ALC49036.1 CH916371 EDV92252.1 ADMH02000875 ETN64797.1 LJIG01016320 KRT80956.1 CH477344 EAT43036.1 AY121679 AAM52006.1 GANO01001473 JAB58398.1 CM000162 EDX03094.1 CH954180 EDV47616.1 L33246 X67202 AF017777 AE014298 JXJN01013884 ATLV01021113 KE525312 KFB46050.1
Proteomes
UP000007151
UP000053240
UP000053268
UP000283053
UP000218220
UP000005204
+ More
UP000007755 UP000078540 UP000005205 UP000242457 UP000005203 UP000235965 UP000027135 UP000002358 UP000053097 UP000000311 UP000009046 UP000078492 UP000076502 UP000078541 UP000078542 UP000192223 UP000053105 UP000279307 UP000007819 UP000069940 UP000249989 UP000001819 UP000007266 UP000008744 UP000268350 UP000095301 UP000009192 UP000095300 UP000192221 UP000001292 UP000091820 UP000008792 UP000007801 UP000076407 UP000037069 UP000075840 UP000007062 UP000092443 UP000078200 UP000092445 UP000075886 UP000092553 UP000076408 UP000001070 UP000069272 UP000000673 UP000075885 UP000008820 UP000075884 UP000002282 UP000008711 UP000000803 UP000092460 UP000075880 UP000030765 UP000075900 UP000075920 UP000075882 UP000075881 UP000075902
UP000007755 UP000078540 UP000005205 UP000242457 UP000005203 UP000235965 UP000027135 UP000002358 UP000053097 UP000000311 UP000009046 UP000078492 UP000076502 UP000078541 UP000078542 UP000192223 UP000053105 UP000279307 UP000007819 UP000069940 UP000249989 UP000001819 UP000007266 UP000008744 UP000268350 UP000095301 UP000009192 UP000095300 UP000192221 UP000001292 UP000091820 UP000008792 UP000007801 UP000076407 UP000037069 UP000075840 UP000007062 UP000092443 UP000078200 UP000092445 UP000075886 UP000092553 UP000076408 UP000001070 UP000069272 UP000000673 UP000075885 UP000008820 UP000075884 UP000002282 UP000008711 UP000000803 UP000092460 UP000075880 UP000030765 UP000075900 UP000075920 UP000075882 UP000075881 UP000075902
Interpro
Gene 3D
ProteinModelPortal
Q1HQB2
A0A212FMM4
A0A194R2R4
A0A194Q015
A0A437B8K6
A0A2A4JXT8
+ More
H9JTV4 F4WRF2 A0A195BE60 A0A158NUL1 A0A2A3EM85 A0A088AKH0 A0A2J7PGQ1 A0A067RHK0 K7J9N2 A0A026W8T3 A0A1S4N210 E2AFV7 E0W3N0 A0A151JQY4 A0A154PRU7 A0A195FJS2 A0A1B6DLZ5 A0A151IFQ3 A0A1W4WZ12 A0A1B6F7U1 A0A0N0U609 A0A0C9Q809 A0A3L8DL59 A0A2S2Q0M9 A0A2H8TPM8 A0A2S2PUK1 A0A1Y1KLI9 W8AZP6 J9K1P5 A0A182H1D8 A0A0A9YUY9 A0A1Q3FWK3 A0A0P4VMD8 A0A0R3NXT6 A0A0K8VHJ0 A0A034VBT2 A0A224XF00 D2A6H6 A0A069DYQ8 A0A0K8S8Y1 A0A0A1WR70 Q29JE4 B4H9X4 A0A0R3NXX1 A0A3B0JQQ1 R4FR01 A0A023F9C1 T1P9J6 B4L5D8 A0A1L8DXH0 A0A1I8Q8V8 A0A1W4UVS1 B4I6H1 A0A1A9W191 B4MAZ6 B3MRW1 A0A182XAK2 A0A0L0BVQ7 A0A182HSE2 Q7QCI4 A0A1I8MCR5 A0A1A9XC23 A0A1A9VB08 A0A0Q9WMC8 A0A1I8Q8T0 A0A2M3ZJH8 A0A1B0A9Y6 A0A2M4ATS3 A0A2M4BSG1 A0A182QGP8 A0A0M5JAL8 A0A182Y6N6 B4JNF9 A0A182FAJ0 W5JK94 A0A1S4FB03 A0A0T6B0R0 A0A182P3T8 Q179W2 A0A182NRB5 Q540V5 U5EUH0 B4PZ33 B3NYD7 P42207 A0A1B0BGK8 A0A182IZ29 A0A084W756 A0A182RQJ2 A0A182W6A5 A0A182W2I2 A0A182LED8 A0A182JP60 A0A182TXN4
H9JTV4 F4WRF2 A0A195BE60 A0A158NUL1 A0A2A3EM85 A0A088AKH0 A0A2J7PGQ1 A0A067RHK0 K7J9N2 A0A026W8T3 A0A1S4N210 E2AFV7 E0W3N0 A0A151JQY4 A0A154PRU7 A0A195FJS2 A0A1B6DLZ5 A0A151IFQ3 A0A1W4WZ12 A0A1B6F7U1 A0A0N0U609 A0A0C9Q809 A0A3L8DL59 A0A2S2Q0M9 A0A2H8TPM8 A0A2S2PUK1 A0A1Y1KLI9 W8AZP6 J9K1P5 A0A182H1D8 A0A0A9YUY9 A0A1Q3FWK3 A0A0P4VMD8 A0A0R3NXT6 A0A0K8VHJ0 A0A034VBT2 A0A224XF00 D2A6H6 A0A069DYQ8 A0A0K8S8Y1 A0A0A1WR70 Q29JE4 B4H9X4 A0A0R3NXX1 A0A3B0JQQ1 R4FR01 A0A023F9C1 T1P9J6 B4L5D8 A0A1L8DXH0 A0A1I8Q8V8 A0A1W4UVS1 B4I6H1 A0A1A9W191 B4MAZ6 B3MRW1 A0A182XAK2 A0A0L0BVQ7 A0A182HSE2 Q7QCI4 A0A1I8MCR5 A0A1A9XC23 A0A1A9VB08 A0A0Q9WMC8 A0A1I8Q8T0 A0A2M3ZJH8 A0A1B0A9Y6 A0A2M4ATS3 A0A2M4BSG1 A0A182QGP8 A0A0M5JAL8 A0A182Y6N6 B4JNF9 A0A182FAJ0 W5JK94 A0A1S4FB03 A0A0T6B0R0 A0A182P3T8 Q179W2 A0A182NRB5 Q540V5 U5EUH0 B4PZ33 B3NYD7 P42207 A0A1B0BGK8 A0A182IZ29 A0A084W756 A0A182RQJ2 A0A182W6A5 A0A182W2I2 A0A182LED8 A0A182JP60 A0A182TXN4
PDB
2QA5
E-value=1.45038e-136,
Score=1245
Ontologies
GO
Topology
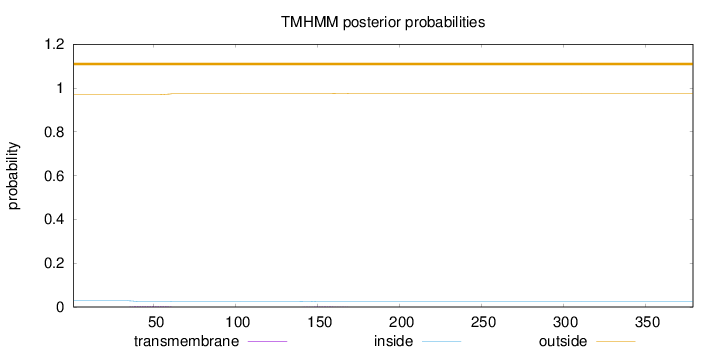
Length:
379
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10231
Exp number, first 60 AAs:
0.07505
Total prob of N-in:
0.02909
outside
1 - 379
Population Genetic Test Statistics
Pi
248.075427
Theta
178.58111
Tajima's D
-1.375433
CLR
0.611347
CSRT
0.0777461126943653
Interpretation
Uncertain
