Pre Gene Modal
BGIBMGA012967
Annotation
hypothetical_protein_KGM_15573_[Danaus_plexippus]
Location in the cell
Nuclear Reliability : 4.174
Sequence
CDS
ATGGCGATAGTAGAAGACGAAATTCAGAACGGTTCTGAAGGGGAAAATGAAATTGAAGTTGTGGTTGAAGAAGATGATGGCGACCAGGATCCAGATTCTGATGATGAAATATTGGAGATGGGAGATTGGCAGAGGAAAGTGGCCGAATATGAACACAAAATCACATTACATCCTTTTAATTATGATGATCACATTGAATTGATCCAGACTTTGTGGCGTCTATCGGAACTGGGGCAATGGCGAACGGCGTTTGAAAGATTATGGGAGATGTCAGTTATAAAAGTTGATCATTGGCTCTTGTGGCTCCGAACAGAAGAGAATATTTCACAGTCTGAAGAACTAATTGCCAATTTATTCTATAAGGCAACCTGTGATTGCTACTCTATGTCTATACTGTCAAGATGGAGTTCGTGGGCCCTTTCATTAGATGAAATACCTAAAATTCGAGCGCAACTTGACGAAGTTTTGCGTCGTGGAGGCTCCGATCCGTATTCAGGAAAAGTGTTGTGGGATCTAAAAATTGACTTTGAAAAATCACAATTAAATGAGACATGCAAAGAAGATTCGGGTTATAAAGAACAAGAAGACCGTGTGTTGAGGTGTTTAGAGGTAGTGGCATCGCTTCCTATAAGCAGAAATCAGGAGGCATGGGATCTATTTGAAGATTTTGCTAAAGATATCCGTGAACAGGAGTACATAGAGAAGGTGGAACAGCAACATAAGGCAGCTGTGGATTACTTAAAGAAAATTATGCCATATGAAATCAGAACAAAGACCACTCAATGTTATATGGCCAATTACAAGGTATACCTTGAGTATATTGATCTTGTGAAGCAGTTGTCTGTAGAAGAGAAGTATGCTGAATGTGATAGCGACGGTCTGCTCAGGCTCCTGTACGACCGGGCGACGTACGAATGTCTGCTCAATCCGGCCGTGTTCGAGTTGCTGGTCGAGTTCGCGAAGCACACCCAGGTGACGTCATCACGCGCGACGTACTGGCGCGTCCTCGAGCTGTGCACGCGGCGCTGTCCGCAGAAGCACACCTTTTGGACCCTCAAGATGCAGCAGGCTGAACACGAGGAGAAAGACTTCGACGAGGTGAAGTCGATCTTCGAGACTGCCTTATCGAAAGGTCTGGAATCTTACAAGGAGGCTGAAATGCTATGGCTGAACTATCTCGAGTACGTGCGCCGCAACACGAAGTTCAGTGACGACGATCAGATTGAGCGGTTGAGGCGCACATTCAGGCTGGCCTGGGACTCGCTTGCTGAGGCATGGGGCGATGAAGCGAATGATTGTGAGATACCATTGTATTGGGCAAGAATTGAATACAAATTCTTGAATTCACCAAAACGAGGGAAAGAGATCTTTGAAGAATTATTCAAATATGGCTTCAATAAAACCATGTCCAAGTATTGGGAAGCTTTGATACAATTAGAAAGTAGCCGGTCTCCTCCGCTGTCCGAGTTCAAGTTCCGGGATCTGCTGAGACGCGCGTTGAGGTCGGTCACCGACTACCCCCCGGCCATAGCGCGCCTCTGGACTGATTACGAACGAGACTACGGGAAGCTGGACACCAGTAAGGAGTGTCAAGAAACTTGTGAGTTAAAAATACGGGAGTGGAGGAAGAATTATCAAGCCTTGAAGGAAAATATGACAAACAAAAACGATACTGATAAAACAAATGATAATAAGAAAGGAAAATCAGCCAACAAAAACAAAAAACTCGAAAACAAACAGAAAGGTAAAAGGAAATTGAATGGCGGTAGTGATGAAGCTAGTGACGTAAAGAGAAAAAGAGACGGAGATCAAGAATTGCCCGTGAAAGAAAAGGACGGAGGTAGCTCTGGAGTCAAGCGGGCACATACTGAAGAAGAGACTGAATTAACTGAGAATAAAAAACAGCGCGTAGAGGCGGCCGGTACAGCGCAGTCTGCGAGCACAGAGGCCTGCACATTGTTCGTCAGTAATCTAGACTTTAAAGTCGATGAAGATAAATTACGACAGGAATTATCGAAGTACGGTGACATCGTCGAAATGCGGCTCAGATCAGGAGTGAAATCGTTCGGCGGCTCTATTTGCTACTGTCAATACAAGACAGCGGACTCAGTGGAGGAGGCTCTGAAGCACGATCGAACTCCTTTAGACGGCAGACCGATGTTCTTGTCTCGATACTCGGCCGATAAATCTAAGGCGACCTTCAAATACGCTACCACGATGGAGAAGAACAAGTTGTTCGTGACGGGCCTGCCTTACTCGCGCTGCAATAAGGAGTGTTTGACCGAAATATTCGAGAAGTACGGCAAACTGAAAGACATCCGGGTGGTCACTTTCAAAGACGGGAAACCGAAAGGTTTGGCGTACGTCGATTACGAGGACGAAGATTCCGCGGCCAGAGCCATAGCGGCGACGAACGGCACGACGGTGGGGGACAGGAAAATAGAGGTCGCGATCAGCCAGCCCCCGCCCCGCGCCGCCCGCCCGACCCCCGCCAGCCGCCCGCCCGCCGCCGGCATGCGGCGGACGCAGCTAAGCTTCCTTCCTCGAGTTTTGCAGCAAGCCTCAACGAGCAAGGAGTCTTCGACAAACAGCCACGCCAACGGCACCGAGAAGCGACCTCTCTCTAATAGTGACTTTAAAAAAATGATTGCGTCATCTTCTTCTTCTCACGACAATTGA
Protein
MAIVEDEIQNGSEGENEIEVVVEEDDGDQDPDSDDEILEMGDWQRKVAEYEHKITLHPFNYDDHIELIQTLWRLSELGQWRTAFERLWEMSVIKVDHWLLWLRTEENISQSEELIANLFYKATCDCYSMSILSRWSSWALSLDEIPKIRAQLDEVLRRGGSDPYSGKVLWDLKIDFEKSQLNETCKEDSGYKEQEDRVLRCLEVVASLPISRNQEAWDLFEDFAKDIREQEYIEKVEQQHKAAVDYLKKIMPYEIRTKTTQCYMANYKVYLEYIDLVKQLSVEEKYAECDSDGLLRLLYDRATYECLLNPAVFELLVEFAKHTQVTSSRATYWRVLELCTRRCPQKHTFWTLKMQQAEHEEKDFDEVKSIFETALSKGLESYKEAEMLWLNYLEYVRRNTKFSDDDQIERLRRTFRLAWDSLAEAWGDEANDCEIPLYWARIEYKFLNSPKRGKEIFEELFKYGFNKTMSKYWEALIQLESSRSPPLSEFKFRDLLRRALRSVTDYPPAIARLWTDYERDYGKLDTSKECQETCELKIREWRKNYQALKENMTNKNDTDKTNDNKKGKSANKNKKLENKQKGKRKLNGGSDEASDVKRKRDGDQELPVKEKDGGSSGVKRAHTEEETELTENKKQRVEAAGTAQSASTEACTLFVSNLDFKVDEDKLRQELSKYGDIVEMRLRSGVKSFGGSICYCQYKTADSVEEALKHDRTPLDGRPMFLSRYSADKSKATFKYATTMEKNKLFVTGLPYSRCNKECLTEIFEKYGKLKDIRVVTFKDGKPKGLAYVDYEDEDSAARAIAATNGTTVGDRKIEVAISQPPPRAARPTPASRPPAAGMRRTQLSFLPRVLQQASTSKESSTNSHANGTEKRPLSNSDFKKMIASSSSSHDN
Summary
Uniprot
H9JTV5
A0A212FMM2
A0A194Q001
A0A194R1Z0
A0A0P6IFE8
A0A0P5SLV3
+ More
A0A0P5S9I1 A0A0N8E0S2 A0A0P6BI09 A0A0C9PZL2 A0A0P4ZG75 A0A2P8YHL8 J9JRG0 A0A2S2PB20 A0A2S2QIL8 A0A182WRR3 A0A182LG21 A0ND71 Q16NI5 A0A182HU15 A0A182VKR9 A0A182NDX2 E2AEC8 A0A182P6M7 A0A182M2X6 A0A2H8U214 A0A182TTA7 A0A1S3F293 A0A1Q3EWF4 A0A182JYB4 A0A182QHB1 A0A2M4BCW7 A0A2M4BCW9 A0A2M3Z7K6 A0A2M4BD54 A0A2M3Z7T4 A0A182FPG4 A0A2M4AE49 A0A0A1XP93 A0A2M4AE37 W5JK77 A0A0A1WXT9 A0A2M4AE83 A0A1J1J8R3 A0A3B4ZYW4 A0A3Q1D6P6 A0A2U4CKK2 A0A3B3CZ66 A0A1S4G755 A0A182RNB7 A0A182VYK3 A0A3Q1B9Z2 H0Z981 A0A182XWC3 A0A2Y9FB44 A0A3Q7QHD7 G1PAD3 A0A3B5A219 U3K2N9 A0A3Q7SKI9 H0WHI7 A0A3L8SCC7 A0A2U4CKK3 A0A0P5ST69 Q16NI3 A0A218V5A0 A0A2K6ATC5 A0A2K5WYB8 F7H2Z6 A0A096N4C5 A0A3Q7QGZ4 A0A2K5YX11 A0A2U3X287 A0A2K5NJF5 A0A091ENF2 A0A340WL08 G1MD89 A0A2Y9F9B7 H2Q6T4 A0A2K5H739 A0A1V4JTC8 A0A3Q7RJK1 A0A2R9C8C5 F7C5B3 A0A2I0M8B6 K9IPK9 E2QXZ2 L5LZJ7 G7PI53 A0A0D9S2R8 H9ERJ3 A0A2U3X280 A0A437D099
A0A0P5S9I1 A0A0N8E0S2 A0A0P6BI09 A0A0C9PZL2 A0A0P4ZG75 A0A2P8YHL8 J9JRG0 A0A2S2PB20 A0A2S2QIL8 A0A182WRR3 A0A182LG21 A0ND71 Q16NI5 A0A182HU15 A0A182VKR9 A0A182NDX2 E2AEC8 A0A182P6M7 A0A182M2X6 A0A2H8U214 A0A182TTA7 A0A1S3F293 A0A1Q3EWF4 A0A182JYB4 A0A182QHB1 A0A2M4BCW7 A0A2M4BCW9 A0A2M3Z7K6 A0A2M4BD54 A0A2M3Z7T4 A0A182FPG4 A0A2M4AE49 A0A0A1XP93 A0A2M4AE37 W5JK77 A0A0A1WXT9 A0A2M4AE83 A0A1J1J8R3 A0A3B4ZYW4 A0A3Q1D6P6 A0A2U4CKK2 A0A3B3CZ66 A0A1S4G755 A0A182RNB7 A0A182VYK3 A0A3Q1B9Z2 H0Z981 A0A182XWC3 A0A2Y9FB44 A0A3Q7QHD7 G1PAD3 A0A3B5A219 U3K2N9 A0A3Q7SKI9 H0WHI7 A0A3L8SCC7 A0A2U4CKK3 A0A0P5ST69 Q16NI3 A0A218V5A0 A0A2K6ATC5 A0A2K5WYB8 F7H2Z6 A0A096N4C5 A0A3Q7QGZ4 A0A2K5YX11 A0A2U3X287 A0A2K5NJF5 A0A091ENF2 A0A340WL08 G1MD89 A0A2Y9F9B7 H2Q6T4 A0A2K5H739 A0A1V4JTC8 A0A3Q7RJK1 A0A2R9C8C5 F7C5B3 A0A2I0M8B6 K9IPK9 E2QXZ2 L5LZJ7 G7PI53 A0A0D9S2R8 H9ERJ3 A0A2U3X280 A0A437D099
Pubmed
EMBL
BABH01003662
AGBW02007651
OWR54940.1
KQ459582
KPI98907.1
KQ460878
+ More
KPJ11539.1 GDIQ01005327 JAN89410.1 GDIP01141636 LRGB01000944 JAL62078.1 KZS14538.1 GDIP01142680 JAL61034.1 GDIQ01073092 JAN21645.1 GDIP01014264 JAM89451.1 GBYB01006998 JAG76765.1 GDIP01213229 JAJ10173.1 PYGN01000588 PSN43747.1 ABLF02015069 ABLF02015070 ABLF02015073 GGMR01013965 MBY26584.1 GGMS01008340 MBY77543.1 AAAB01008859 EAU77058.3 CH477823 EAT35909.1 APCN01000599 GL438827 EFN68361.1 AXCM01005292 GFXV01007713 MBW19518.1 GFDL01015410 JAV19635.1 AXCN02001589 GGFJ01001749 MBW50890.1 GGFJ01001748 MBW50889.1 GGFM01003743 MBW24494.1 GGFJ01001750 MBW50891.1 GGFM01003747 MBW24498.1 GGFK01005740 MBW39061.1 GBXI01001108 JAD13184.1 GGFK01005732 MBW39053.1 ADMH02001267 ETN63295.1 GBXI01010423 JAD03869.1 GGFK01005739 MBW39060.1 CVRI01000070 CRL07305.1 ABQF01028936 ABQF01028937 ABQF01028938 AAPE02036205 AGTO01020434 AAQR03070029 AAQR03070030 AAQR03070031 AAQR03070032 AAQR03070033 QUSF01000030 RLV99915.1 GDIP01135628 JAL68086.1 EAT35911.1 MUZQ01000045 OWK61146.1 AQIA01012596 AQIA01012597 AQIA01012598 JSUE03007704 AHZZ02002934 KK718897 KFO59463.1 ACTA01195906 AACZ04070370 LSYS01006629 OPJ74967.1 AJFE02116941 AJFE02116942 AKCR02000030 PKK25915.1 GABZ01004088 JAA49437.1 AAEX03014754 KB106121 ELK31467.1 CM001286 EHH66680.1 AQIB01025165 AQIB01025166 AQIB01025167 JU321246 JV048821 CM001259 AFE65002.1 AFI38892.1 EHH27376.1 CM012445 RVE68294.1
KPJ11539.1 GDIQ01005327 JAN89410.1 GDIP01141636 LRGB01000944 JAL62078.1 KZS14538.1 GDIP01142680 JAL61034.1 GDIQ01073092 JAN21645.1 GDIP01014264 JAM89451.1 GBYB01006998 JAG76765.1 GDIP01213229 JAJ10173.1 PYGN01000588 PSN43747.1 ABLF02015069 ABLF02015070 ABLF02015073 GGMR01013965 MBY26584.1 GGMS01008340 MBY77543.1 AAAB01008859 EAU77058.3 CH477823 EAT35909.1 APCN01000599 GL438827 EFN68361.1 AXCM01005292 GFXV01007713 MBW19518.1 GFDL01015410 JAV19635.1 AXCN02001589 GGFJ01001749 MBW50890.1 GGFJ01001748 MBW50889.1 GGFM01003743 MBW24494.1 GGFJ01001750 MBW50891.1 GGFM01003747 MBW24498.1 GGFK01005740 MBW39061.1 GBXI01001108 JAD13184.1 GGFK01005732 MBW39053.1 ADMH02001267 ETN63295.1 GBXI01010423 JAD03869.1 GGFK01005739 MBW39060.1 CVRI01000070 CRL07305.1 ABQF01028936 ABQF01028937 ABQF01028938 AAPE02036205 AGTO01020434 AAQR03070029 AAQR03070030 AAQR03070031 AAQR03070032 AAQR03070033 QUSF01000030 RLV99915.1 GDIP01135628 JAL68086.1 EAT35911.1 MUZQ01000045 OWK61146.1 AQIA01012596 AQIA01012597 AQIA01012598 JSUE03007704 AHZZ02002934 KK718897 KFO59463.1 ACTA01195906 AACZ04070370 LSYS01006629 OPJ74967.1 AJFE02116941 AJFE02116942 AKCR02000030 PKK25915.1 GABZ01004088 JAA49437.1 AAEX03014754 KB106121 ELK31467.1 CM001286 EHH66680.1 AQIB01025165 AQIB01025166 AQIB01025167 JU321246 JV048821 CM001259 AFE65002.1 AFI38892.1 EHH27376.1 CM012445 RVE68294.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000076858
UP000245037
+ More
UP000007819 UP000076407 UP000075882 UP000007062 UP000008820 UP000075840 UP000075903 UP000075884 UP000000311 UP000075885 UP000075883 UP000075902 UP000081671 UP000075881 UP000075886 UP000069272 UP000000673 UP000183832 UP000261400 UP000257160 UP000245320 UP000261560 UP000075900 UP000075920 UP000007754 UP000076408 UP000248484 UP000286641 UP000001074 UP000016665 UP000286640 UP000005225 UP000276834 UP000197619 UP000233120 UP000233100 UP000006718 UP000028761 UP000233140 UP000245340 UP000233060 UP000052976 UP000265300 UP000008912 UP000002277 UP000233080 UP000190648 UP000240080 UP000008225 UP000053872 UP000002254 UP000009130 UP000029965
UP000007819 UP000076407 UP000075882 UP000007062 UP000008820 UP000075840 UP000075903 UP000075884 UP000000311 UP000075885 UP000075883 UP000075902 UP000081671 UP000075881 UP000075886 UP000069272 UP000000673 UP000183832 UP000261400 UP000257160 UP000245320 UP000261560 UP000075900 UP000075920 UP000007754 UP000076408 UP000248484 UP000286641 UP000001074 UP000016665 UP000286640 UP000005225 UP000276834 UP000197619 UP000233120 UP000233100 UP000006718 UP000028761 UP000233140 UP000245340 UP000233060 UP000052976 UP000265300 UP000008912 UP000002277 UP000233080 UP000190648 UP000240080 UP000008225 UP000053872 UP000002254 UP000009130 UP000029965
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JTV5
A0A212FMM2
A0A194Q001
A0A194R1Z0
A0A0P6IFE8
A0A0P5SLV3
+ More
A0A0P5S9I1 A0A0N8E0S2 A0A0P6BI09 A0A0C9PZL2 A0A0P4ZG75 A0A2P8YHL8 J9JRG0 A0A2S2PB20 A0A2S2QIL8 A0A182WRR3 A0A182LG21 A0ND71 Q16NI5 A0A182HU15 A0A182VKR9 A0A182NDX2 E2AEC8 A0A182P6M7 A0A182M2X6 A0A2H8U214 A0A182TTA7 A0A1S3F293 A0A1Q3EWF4 A0A182JYB4 A0A182QHB1 A0A2M4BCW7 A0A2M4BCW9 A0A2M3Z7K6 A0A2M4BD54 A0A2M3Z7T4 A0A182FPG4 A0A2M4AE49 A0A0A1XP93 A0A2M4AE37 W5JK77 A0A0A1WXT9 A0A2M4AE83 A0A1J1J8R3 A0A3B4ZYW4 A0A3Q1D6P6 A0A2U4CKK2 A0A3B3CZ66 A0A1S4G755 A0A182RNB7 A0A182VYK3 A0A3Q1B9Z2 H0Z981 A0A182XWC3 A0A2Y9FB44 A0A3Q7QHD7 G1PAD3 A0A3B5A219 U3K2N9 A0A3Q7SKI9 H0WHI7 A0A3L8SCC7 A0A2U4CKK3 A0A0P5ST69 Q16NI3 A0A218V5A0 A0A2K6ATC5 A0A2K5WYB8 F7H2Z6 A0A096N4C5 A0A3Q7QGZ4 A0A2K5YX11 A0A2U3X287 A0A2K5NJF5 A0A091ENF2 A0A340WL08 G1MD89 A0A2Y9F9B7 H2Q6T4 A0A2K5H739 A0A1V4JTC8 A0A3Q7RJK1 A0A2R9C8C5 F7C5B3 A0A2I0M8B6 K9IPK9 E2QXZ2 L5LZJ7 G7PI53 A0A0D9S2R8 H9ERJ3 A0A2U3X280 A0A437D099
A0A0P5S9I1 A0A0N8E0S2 A0A0P6BI09 A0A0C9PZL2 A0A0P4ZG75 A0A2P8YHL8 J9JRG0 A0A2S2PB20 A0A2S2QIL8 A0A182WRR3 A0A182LG21 A0ND71 Q16NI5 A0A182HU15 A0A182VKR9 A0A182NDX2 E2AEC8 A0A182P6M7 A0A182M2X6 A0A2H8U214 A0A182TTA7 A0A1S3F293 A0A1Q3EWF4 A0A182JYB4 A0A182QHB1 A0A2M4BCW7 A0A2M4BCW9 A0A2M3Z7K6 A0A2M4BD54 A0A2M3Z7T4 A0A182FPG4 A0A2M4AE49 A0A0A1XP93 A0A2M4AE37 W5JK77 A0A0A1WXT9 A0A2M4AE83 A0A1J1J8R3 A0A3B4ZYW4 A0A3Q1D6P6 A0A2U4CKK2 A0A3B3CZ66 A0A1S4G755 A0A182RNB7 A0A182VYK3 A0A3Q1B9Z2 H0Z981 A0A182XWC3 A0A2Y9FB44 A0A3Q7QHD7 G1PAD3 A0A3B5A219 U3K2N9 A0A3Q7SKI9 H0WHI7 A0A3L8SCC7 A0A2U4CKK3 A0A0P5ST69 Q16NI3 A0A218V5A0 A0A2K6ATC5 A0A2K5WYB8 F7H2Z6 A0A096N4C5 A0A3Q7QGZ4 A0A2K5YX11 A0A2U3X287 A0A2K5NJF5 A0A091ENF2 A0A340WL08 G1MD89 A0A2Y9F9B7 H2Q6T4 A0A2K5H739 A0A1V4JTC8 A0A3Q7RJK1 A0A2R9C8C5 F7C5B3 A0A2I0M8B6 K9IPK9 E2QXZ2 L5LZJ7 G7PI53 A0A0D9S2R8 H9ERJ3 A0A2U3X280 A0A437D099
PDB
5JPZ
E-value=3.74384e-23,
Score=271
Ontologies
GO
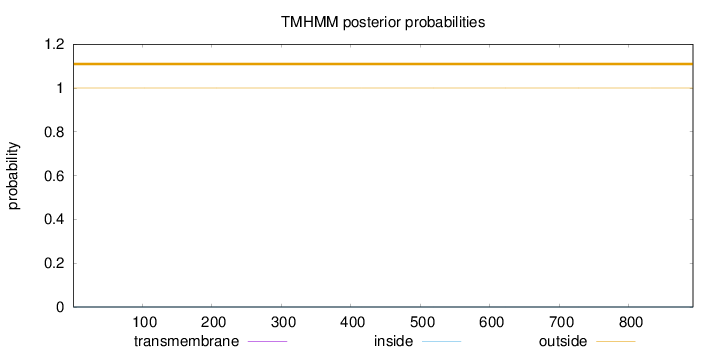
Topology
Length:
892
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0004
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00004
outside
1 - 892
Population Genetic Test Statistics
Pi
171.078048
Theta
164.72258
Tajima's D
0.01467
CLR
340.840075
CSRT
0.371481425928704
Interpretation
Uncertain
