Gene
KWMTBOMO09655
Pre Gene Modal
BGIBMGA012969
Annotation
PREDICTED:_exocyst_complex_component_8_[Papilio_machaon]
Full name
Proteasome endopeptidase complex
Location in the cell
Nuclear Reliability : 2.976
Sequence
CDS
ATGTCCGAGATAGATATGGATAAGTTTGCTGTCCCAGACTTCGTCCCCGAACGTTACGTAAAAGACCTCGCTAGATCCTGCGTCGGAGGGGAGGAGCTACAGCAACAGAAGGAGAAAATACAAAATTTAGCCGAAGAAACCGCAAGTGCCCTAAAGAAAAATGTCTACGAGAATTATATGCAGTTTATCGAGACTGCGACGGAGATATCCCACCTCGAAACAGAAATGTACAAGCTATCGCATTTGTTAAGCGAACAGCGTTCTGTGCTGACTGTGCTCTCCCACGCCTCAATCCTCGGTAACGAGAACGGTTTATCACGCGAGTCTCTTGATAATCACGACGTCGAGGCTGAGAGGGCAGAAGAGGAACGAAAAAATAAATTAAATCTCATTACAGAAAAGGTAGAATCATGCATGAATCTGTTAGATGTCCCAGACCGAACTCTGCTTCATGAAGGTGACTTGCTAGAAATAGATGCAGAAGAAAATACTGCATTACAAAGAATGCATGTATATTTATTTAATGACTGCCTTATGTTAACATCCTGGATTTCTAACAGACGTGGTCCAATGAGATATAAGTTTCAAGTGAGTTACCCAATAAAGAATTTAGCAGTGGTGAATGTAAGAGATTTAGGAACTGTGAAACAAGCATTTAAAGTGTTAGCCTTCCCAGACACCAGAGTATTTCAATGCAATAGTTTTGCGATAAAAAAAGATTGGCTAGACAAATTTGAGATAGCAAAAAAACTGTATATTGAAAAAGAAAATTCTAAACAGAAGAAAAAAGATAGTCAAGAAAAATCAAAATCTGAATCTATTGAATCACCGAGTCTGTCTGTGGAAGAGATGCCATCACCCCAAATTACACCATTGCCTGAGTGGTTGGCTGATGTTACTGAAGAGCTTGATGTTTTGATTGTGGAGAGACATTTTCAAAAGACCTATGAGTTGATGGTTTCAGCTCAAAAAGCTTTAAATACTGAAGATTTTAAAAACCATCCACAAAGGGTTGATATATTGAAAAAAATAGAACAGAAGCAAAAGAGTTTAATTGAAATATTATTGAAAGATTTAGATGTAACACATAATTGGAGTTTGCGAGGAGGTCTCCAATCAACTAGACAACCGGTACTGATACTCAATAAGTTTAATATGACCAGTCAGGCTTGTGACTTGTTTTTGACGATATGCAGTTACACAGTCAAAGTTCATGTGAAAAGGGTGCAACTGGAAGGGTCAATTAACACTTATGTCAAACAAGTGGGTGAAGTGTTTTTTTGCTCATTGAGTGATGCCTTAACAGAGTTTATTTCACTATTTCCTAAAAATAAAATAAGTGCTTTTATAGTCTGGGCTAGTCTGCAACTTAGGATATTGATGAGTCAGATTATAAAACAAGTGTTCACACCTCAGTGTGCATTAGATTCCGTTCAGGATTGTGTTGAGAGTCTAAGGGAACAATGTACATTGTTTTGTCAATTTGGTTTAGATTTAAGATTCCAGTTGAACAGTTGTTTGAGAACACCAATAATAAAAGCTTTGAGAGAATATAAAGAGAAGACTGTTGACTCTATGAAAAATAAAATCACTGAAGACAAATGGACTCCAGTGAATATGCACACCAAAGCAGGTGTCAATAAATTCCTGAAACAAATGGAAACTTTGGGCCTTATATTAGCTAAGTATGTAATTAATGAAACATGGGTAGATTTATCAAGTAGTACCATATGGTTTACAAAGTCTATTGTCACTATACAGAAAATAGGTCTCCAGTTAGTGAACAAAGATATGATGGATAACCTTGATGAATACATTTATACAATATTCAATTCATGGCTTGTCCTCGCATCAGTCAACGATATTAGTTATGCAAAGAAGAATTTCAAATTCATTTTAGAAACAGTGATGCCTTTAATGCTTAGATGTTATAAAGAAGAAGTTGGCTATGATAATGAGAAATTAGTAGCTTTGGCAAAGAACTATGGTGTCAGTATTACACCATCCAAAAACATCACAAAATACACAAGTAATGAATATTTATGA
Protein
MSEIDMDKFAVPDFVPERYVKDLARSCVGGEELQQQKEKIQNLAEETASALKKNVYENYMQFIETATEISHLETEMYKLSHLLSEQRSVLTVLSHASILGNENGLSRESLDNHDVEAERAEEERKNKLNLITEKVESCMNLLDVPDRTLLHEGDLLEIDAEENTALQRMHVYLFNDCLMLTSWISNRRGPMRYKFQVSYPIKNLAVVNVRDLGTVKQAFKVLAFPDTRVFQCNSFAIKKDWLDKFEIAKKLYIEKENSKQKKKDSQEKSKSESIESPSLSVEEMPSPQITPLPEWLADVTEELDVLIVERHFQKTYELMVSAQKALNTEDFKNHPQRVDILKKIEQKQKSLIEILLKDLDVTHNWSLRGGLQSTRQPVLILNKFNMTSQACDLFLTICSYTVKVHVKRVQLEGSINTYVKQVGEVFFCSLSDALTEFISLFPKNKISAFIVWASLQLRILMSQIIKQVFTPQCALDSVQDCVESLREQCTLFCQFGLDLRFQLNSCLRTPIIKALREYKEKTVDSMKNKITEDKWTPVNMHTKAGVNKFLKQMETLGLILAKYVINETWVDLSSSTIWFTKSIVTIQKIGLQLVNKDMMDNLDEYIYTIFNSWLVLASVNDISYAKKNFKFILETVMPLMLRCYKEEVGYDNEKLVALAKNYGVSITPSKNITKYTSNEYL
Summary
Catalytic Activity
Cleavage of peptide bonds with very broad specificity.
Similarity
Belongs to the peptidase T1A family.
Uniprot
H9JTV7
A0A2A4JWS7
A0A437BAM3
A0A2H1VMV5
A0A194R6P5
A0A194Q674
+ More
A0A0L7LCI5 A0A1E1WQS5 A0A212FMI3 A0A067QXD9 A0A2J7QKH9 A0A2J7QKI3 A0A2J7QKK5 A0A1W4WPP4 A0A1Y1KF15 A0A1B6FB48 A0A1B6DF58 D6WJN8 A0A154PRT0 A0A0L7RA92 N6TBX2 A0A069DWT4 A0A087ZNT1 A0A026WLH8 A0A3L8DAM5 T1I6U3 A0A0V0G7K9 A0A0M9A0A9 E9IBT5 A0A158NKD1 E2C1N0 A0A310SM70 E0VFG0 E2A3M3 A0A0J7KSV3 A0A2H8TVE6 J9JVA0 A0A195EIQ0 A0A1B6HHE7 A0A0T6B6E5 A0A0A9YHQ1 A0A2A3ENH6 A0A151I9F5 E9GF51 A0A0P5G0N6 A0A0P5UQQ0 A0A0P5ZNR5 A0A0P5NV06 A0A1B6KMY6 A0A0N8BAI8 A0A0P6EBH1 A0A0P6CQB9 A0A0P6GQC8 A0A0P6CL20 A0A0P6G4W3 A0A0P6G3J7 A0A0N8CCX2 A0A0P6CTG3 A0A1S3JCI7 A0A0P5ZGU2 A0A0P4X6Y4 A0A0N8EID2 A0A0N8EBX6 F4W8N0 A0A0N8CU89 A0A195B0R5 A0A0P4WL32 A0A195FBF9 A0A151WSZ9 A0A1S3HX88 A0A3P8SUE2 A0A3Q1BGS1 A0A3B5BL39 A0A3Q0RKG9 T1J4C7 A0A3Q1F4G1 H2MTA2 A0A3Q4H905 A0A3P8RFW8 A0A3P9BCM2 W5LTD5 W5NN81 A0A3B3H6H8 I3KPK8 A0A3Q3NFG5 A0A3B4WTG8 A0A3B4TM95 A0A3B3ZJA1 A0A3B4H3J6 A0A3Q2VM53 A0A3Q3N4T2 A4IG64 A0A3N0YKB1 F1QE13 A0A210PXP0 A0A3P9JJF3 A0A3B4CYR8
A0A0L7LCI5 A0A1E1WQS5 A0A212FMI3 A0A067QXD9 A0A2J7QKH9 A0A2J7QKI3 A0A2J7QKK5 A0A1W4WPP4 A0A1Y1KF15 A0A1B6FB48 A0A1B6DF58 D6WJN8 A0A154PRT0 A0A0L7RA92 N6TBX2 A0A069DWT4 A0A087ZNT1 A0A026WLH8 A0A3L8DAM5 T1I6U3 A0A0V0G7K9 A0A0M9A0A9 E9IBT5 A0A158NKD1 E2C1N0 A0A310SM70 E0VFG0 E2A3M3 A0A0J7KSV3 A0A2H8TVE6 J9JVA0 A0A195EIQ0 A0A1B6HHE7 A0A0T6B6E5 A0A0A9YHQ1 A0A2A3ENH6 A0A151I9F5 E9GF51 A0A0P5G0N6 A0A0P5UQQ0 A0A0P5ZNR5 A0A0P5NV06 A0A1B6KMY6 A0A0N8BAI8 A0A0P6EBH1 A0A0P6CQB9 A0A0P6GQC8 A0A0P6CL20 A0A0P6G4W3 A0A0P6G3J7 A0A0N8CCX2 A0A0P6CTG3 A0A1S3JCI7 A0A0P5ZGU2 A0A0P4X6Y4 A0A0N8EID2 A0A0N8EBX6 F4W8N0 A0A0N8CU89 A0A195B0R5 A0A0P4WL32 A0A195FBF9 A0A151WSZ9 A0A1S3HX88 A0A3P8SUE2 A0A3Q1BGS1 A0A3B5BL39 A0A3Q0RKG9 T1J4C7 A0A3Q1F4G1 H2MTA2 A0A3Q4H905 A0A3P8RFW8 A0A3P9BCM2 W5LTD5 W5NN81 A0A3B3H6H8 I3KPK8 A0A3Q3NFG5 A0A3B4WTG8 A0A3B4TM95 A0A3B3ZJA1 A0A3B4H3J6 A0A3Q2VM53 A0A3Q3N4T2 A4IG64 A0A3N0YKB1 F1QE13 A0A210PXP0 A0A3P9JJF3 A0A3B4CYR8
EC Number
3.4.25.1
Pubmed
EMBL
BABH01003667
NWSH01000463
PCG76236.1
RSAL01000102
RVE47457.1
ODYU01003407
+ More
SOQ42137.1 KQ460878 KPJ11536.1 KQ459582 KPI98910.1 JTDY01001675 KOB73188.1 GDQN01001712 JAT89342.1 AGBW02007651 OWR54943.1 KK852902 KDR14056.1 NEVH01013262 PNF29095.1 PNF29096.1 PNF29098.1 GEZM01085213 JAV60113.1 GECZ01022357 JAS47412.1 GEDC01012991 JAS24307.1 KQ971343 EFA04473.2 KQ435090 KZC14612.1 KQ414619 KOC67764.1 APGK01043869 KB741018 KB632152 ENN75228.1 ERL89141.1 GBGD01000722 JAC88167.1 KK107182 EZA55954.1 QOIP01000011 RLU17183.1 ACPB03000880 GECL01001994 JAP04130.1 KQ435784 KOX74557.1 GL762137 EFZ21925.1 ADTU01018781 GL451937 EFN78206.1 KQ761343 OAD57875.1 DS235111 EEB12116.1 GL436457 EFN71915.1 LBMM01003526 KMQ93418.1 GFXV01005727 MBW17532.1 ABLF02008446 KQ978881 KYN27739.1 GECU01033695 JAS74011.1 LJIG01009506 KRT82943.1 GBHO01012997 GBHO01012996 GBRD01004489 JAG30607.1 JAG30608.1 JAG61332.1 KZ288204 PBC33315.1 KQ978323 KYM95116.1 GL732541 EFX81943.1 GDIQ01248376 GDIP01064955 JAK03349.1 JAM38760.1 GDIP01109690 JAL94024.1 GDIP01044798 JAM58917.1 GDIQ01147464 JAL04262.1 GEBQ01027158 GEBQ01000003 JAT12819.1 JAT39974.1 GDIQ01196755 JAK54970.1 GDIQ01077186 JAN17551.1 GDIQ01089634 JAN05103.1 GDIQ01030259 JAN64478.1 GDIQ01089633 JAN05104.1 GDIQ01049366 JAN45371.1 GDIQ01038963 JAN55774.1 GDIQ01092083 JAL59643.1 GDIP01009426 JAM94289.1 GDIP01044797 LRGB01000248 JAM58918.1 KZS20069.1 GDIP01253719 JAI69682.1 GDIQ01023812 JAN70925.1 GDIQ01041860 JAN52877.1 GL887908 EGI69521.1 GDIP01098335 JAM05380.1 KQ976692 KYM77794.1 GDRN01042105 JAI67071.1 KQ981693 KYN37711.1 KQ982762 KYQ51029.1 JH431842 AHAT01008283 AERX01042016 BC134948 AAI34949.1 RJVU01038599 ROL46291.1 CABZ01042340 NEDP02005415 OWF41251.1
SOQ42137.1 KQ460878 KPJ11536.1 KQ459582 KPI98910.1 JTDY01001675 KOB73188.1 GDQN01001712 JAT89342.1 AGBW02007651 OWR54943.1 KK852902 KDR14056.1 NEVH01013262 PNF29095.1 PNF29096.1 PNF29098.1 GEZM01085213 JAV60113.1 GECZ01022357 JAS47412.1 GEDC01012991 JAS24307.1 KQ971343 EFA04473.2 KQ435090 KZC14612.1 KQ414619 KOC67764.1 APGK01043869 KB741018 KB632152 ENN75228.1 ERL89141.1 GBGD01000722 JAC88167.1 KK107182 EZA55954.1 QOIP01000011 RLU17183.1 ACPB03000880 GECL01001994 JAP04130.1 KQ435784 KOX74557.1 GL762137 EFZ21925.1 ADTU01018781 GL451937 EFN78206.1 KQ761343 OAD57875.1 DS235111 EEB12116.1 GL436457 EFN71915.1 LBMM01003526 KMQ93418.1 GFXV01005727 MBW17532.1 ABLF02008446 KQ978881 KYN27739.1 GECU01033695 JAS74011.1 LJIG01009506 KRT82943.1 GBHO01012997 GBHO01012996 GBRD01004489 JAG30607.1 JAG30608.1 JAG61332.1 KZ288204 PBC33315.1 KQ978323 KYM95116.1 GL732541 EFX81943.1 GDIQ01248376 GDIP01064955 JAK03349.1 JAM38760.1 GDIP01109690 JAL94024.1 GDIP01044798 JAM58917.1 GDIQ01147464 JAL04262.1 GEBQ01027158 GEBQ01000003 JAT12819.1 JAT39974.1 GDIQ01196755 JAK54970.1 GDIQ01077186 JAN17551.1 GDIQ01089634 JAN05103.1 GDIQ01030259 JAN64478.1 GDIQ01089633 JAN05104.1 GDIQ01049366 JAN45371.1 GDIQ01038963 JAN55774.1 GDIQ01092083 JAL59643.1 GDIP01009426 JAM94289.1 GDIP01044797 LRGB01000248 JAM58918.1 KZS20069.1 GDIP01253719 JAI69682.1 GDIQ01023812 JAN70925.1 GDIQ01041860 JAN52877.1 GL887908 EGI69521.1 GDIP01098335 JAM05380.1 KQ976692 KYM77794.1 GDRN01042105 JAI67071.1 KQ981693 KYN37711.1 KQ982762 KYQ51029.1 JH431842 AHAT01008283 AERX01042016 BC134948 AAI34949.1 RJVU01038599 ROL46291.1 CABZ01042340 NEDP02005415 OWF41251.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000037510
+ More
UP000007151 UP000027135 UP000235965 UP000192223 UP000007266 UP000076502 UP000053825 UP000019118 UP000030742 UP000005203 UP000053097 UP000279307 UP000015103 UP000053105 UP000005205 UP000008237 UP000009046 UP000000311 UP000036403 UP000007819 UP000078492 UP000242457 UP000078542 UP000000305 UP000085678 UP000076858 UP000007755 UP000078540 UP000078541 UP000075809 UP000265080 UP000257160 UP000261400 UP000261340 UP000257200 UP000001038 UP000261580 UP000265100 UP000265160 UP000018467 UP000018468 UP000005207 UP000261660 UP000261360 UP000261420 UP000261520 UP000261460 UP000264840 UP000261640 UP000000437 UP000242188 UP000265200 UP000261440
UP000007151 UP000027135 UP000235965 UP000192223 UP000007266 UP000076502 UP000053825 UP000019118 UP000030742 UP000005203 UP000053097 UP000279307 UP000015103 UP000053105 UP000005205 UP000008237 UP000009046 UP000000311 UP000036403 UP000007819 UP000078492 UP000242457 UP000078542 UP000000305 UP000085678 UP000076858 UP000007755 UP000078540 UP000078541 UP000075809 UP000265080 UP000257160 UP000261400 UP000261340 UP000257200 UP000001038 UP000261580 UP000265100 UP000265160 UP000018467 UP000018468 UP000005207 UP000261660 UP000261360 UP000261420 UP000261520 UP000261460 UP000264840 UP000261640 UP000000437 UP000242188 UP000265200 UP000261440
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JTV7
A0A2A4JWS7
A0A437BAM3
A0A2H1VMV5
A0A194R6P5
A0A194Q674
+ More
A0A0L7LCI5 A0A1E1WQS5 A0A212FMI3 A0A067QXD9 A0A2J7QKH9 A0A2J7QKI3 A0A2J7QKK5 A0A1W4WPP4 A0A1Y1KF15 A0A1B6FB48 A0A1B6DF58 D6WJN8 A0A154PRT0 A0A0L7RA92 N6TBX2 A0A069DWT4 A0A087ZNT1 A0A026WLH8 A0A3L8DAM5 T1I6U3 A0A0V0G7K9 A0A0M9A0A9 E9IBT5 A0A158NKD1 E2C1N0 A0A310SM70 E0VFG0 E2A3M3 A0A0J7KSV3 A0A2H8TVE6 J9JVA0 A0A195EIQ0 A0A1B6HHE7 A0A0T6B6E5 A0A0A9YHQ1 A0A2A3ENH6 A0A151I9F5 E9GF51 A0A0P5G0N6 A0A0P5UQQ0 A0A0P5ZNR5 A0A0P5NV06 A0A1B6KMY6 A0A0N8BAI8 A0A0P6EBH1 A0A0P6CQB9 A0A0P6GQC8 A0A0P6CL20 A0A0P6G4W3 A0A0P6G3J7 A0A0N8CCX2 A0A0P6CTG3 A0A1S3JCI7 A0A0P5ZGU2 A0A0P4X6Y4 A0A0N8EID2 A0A0N8EBX6 F4W8N0 A0A0N8CU89 A0A195B0R5 A0A0P4WL32 A0A195FBF9 A0A151WSZ9 A0A1S3HX88 A0A3P8SUE2 A0A3Q1BGS1 A0A3B5BL39 A0A3Q0RKG9 T1J4C7 A0A3Q1F4G1 H2MTA2 A0A3Q4H905 A0A3P8RFW8 A0A3P9BCM2 W5LTD5 W5NN81 A0A3B3H6H8 I3KPK8 A0A3Q3NFG5 A0A3B4WTG8 A0A3B4TM95 A0A3B3ZJA1 A0A3B4H3J6 A0A3Q2VM53 A0A3Q3N4T2 A4IG64 A0A3N0YKB1 F1QE13 A0A210PXP0 A0A3P9JJF3 A0A3B4CYR8
A0A0L7LCI5 A0A1E1WQS5 A0A212FMI3 A0A067QXD9 A0A2J7QKH9 A0A2J7QKI3 A0A2J7QKK5 A0A1W4WPP4 A0A1Y1KF15 A0A1B6FB48 A0A1B6DF58 D6WJN8 A0A154PRT0 A0A0L7RA92 N6TBX2 A0A069DWT4 A0A087ZNT1 A0A026WLH8 A0A3L8DAM5 T1I6U3 A0A0V0G7K9 A0A0M9A0A9 E9IBT5 A0A158NKD1 E2C1N0 A0A310SM70 E0VFG0 E2A3M3 A0A0J7KSV3 A0A2H8TVE6 J9JVA0 A0A195EIQ0 A0A1B6HHE7 A0A0T6B6E5 A0A0A9YHQ1 A0A2A3ENH6 A0A151I9F5 E9GF51 A0A0P5G0N6 A0A0P5UQQ0 A0A0P5ZNR5 A0A0P5NV06 A0A1B6KMY6 A0A0N8BAI8 A0A0P6EBH1 A0A0P6CQB9 A0A0P6GQC8 A0A0P6CL20 A0A0P6G4W3 A0A0P6G3J7 A0A0N8CCX2 A0A0P6CTG3 A0A1S3JCI7 A0A0P5ZGU2 A0A0P4X6Y4 A0A0N8EID2 A0A0N8EBX6 F4W8N0 A0A0N8CU89 A0A195B0R5 A0A0P4WL32 A0A195FBF9 A0A151WSZ9 A0A1S3HX88 A0A3P8SUE2 A0A3Q1BGS1 A0A3B5BL39 A0A3Q0RKG9 T1J4C7 A0A3Q1F4G1 H2MTA2 A0A3Q4H905 A0A3P8RFW8 A0A3P9BCM2 W5LTD5 W5NN81 A0A3B3H6H8 I3KPK8 A0A3Q3NFG5 A0A3B4WTG8 A0A3B4TM95 A0A3B3ZJA1 A0A3B4H3J6 A0A3Q2VM53 A0A3Q3N4T2 A4IG64 A0A3N0YKB1 F1QE13 A0A210PXP0 A0A3P9JJF3 A0A3B4CYR8
PDB
1ZC3
E-value=2.40939e-21,
Score=254
Ontologies
PANTHER
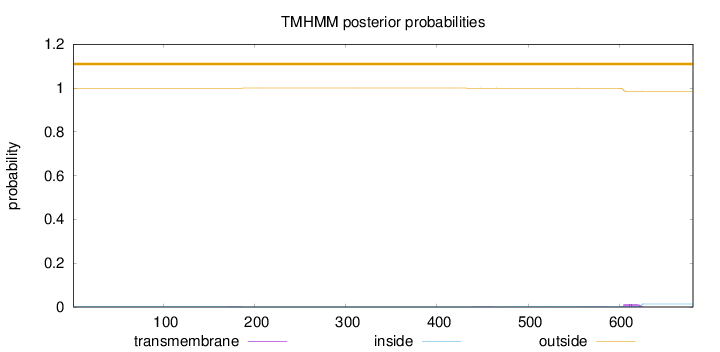
Topology
Length:
681
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.28386
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00059
outside
1 - 681
Population Genetic Test Statistics
Pi
212.128496
Theta
178.700372
Tajima's D
1.116735
CLR
0
CSRT
0.688815559222039
Interpretation
Uncertain
