Gene
KWMTBOMO09653
Pre Gene Modal
BGIBMGA012971
Annotation
PREDICTED:_small_G_protein_signaling_modulator_2-like_isoform_X4_[Papilio_polytes]
Full name
ATP-dependent DNA helicase
Location in the cell
Nuclear Reliability : 2.464
Sequence
CDS
ATGCCGGCAGCTAATGCCCCCACGGACCCTGAAACAGAGCAGAAAGAACGACTTATAGCGGCTGTGAAGAAGGAAGTCAAGCAGCTGATGGAAGAGGCTGTTACGAGGAAATTTGTCCATGAAGATAGTGGTGGAGTGACAACGTTGTGTGGCGCCGTTGAAGCGTGCCTGGGTCAAGGGTTGCGACGAAGAGCTCTCGGTCTATTTAAGACCAGTTCAACGACAGCTTTGCTGCACAAAATTGCAAAGCATTGCCCCGAAGCTGCATTAGTATCAGCCCGGGTGCTTGCTGCCGAAGGAACGCCGGCAACAAGGTCCGCTTCCGGAGTGGAGAGGCGTCCATCAGCGCCACGACCTCCTCTTTGTAAGAGAGGAAGCGGTTCTCTACTCGCGCAATCTCCGCCACAGTCTAAATACCTCTGGATTCGTATAGCTCTATTCGAGAAGCAGCTGGCAAAAATCATTGAGCATTTAGTGACGAATGCTACTCGTTATTATGAACGTGATGCTCTAGTGGCCGACCCTGACTACGGCAGTATTCTTAGTTCACTGCTAGTAGGGCCATGCGCGCTAGAATATTCAAAGGCTAAACCACCGGTTACATTCTGGACCGATCCACCAGCTGATGAATTGGTTCAAAGACATCGTATGTCAGCGGGTACAACGACACCGCCTTCAGTTCGAAGACCGATTTTGAACTTTAAAAGGAGTCTAAATGCCTCGTCTGACGATAGTTCGTCAGTCAACACAGTTACGGGCAAGGGAACGACAAGTGCCAAAGATTACGTAGAAAGTTTACATCAGAATTCAACGGCTACGCTTCTATACGGAAAGAACAATGTTCGCGTACAGCCTAAAGACGCAGAGGAACCCATGGCAGGATATTTGAGCCTTCACCAGACGGCGGTTGGATTGGTGATTAAATGGACACCAAATCAGCTCATGAATGGTTACGCCGAAAGCGAGAGTGTTGACAAAAGTGTGTACTGGGCGATGTCACTGCAAGTGCAAGTGTGGGAGGTGGTATACGTGCACGTGCATCGGTCTCAAGGCAGCGACGCGCTCATACTTGTCGGCCAGGACGGAGTGCAGAGACCACCCATACACTTCCCGAAAGGAGGTCACTTGCTGTCTTTTTTGAGCAATCTAGAGACAGGTTTATTACCTCATGGTCAGCTAGATCCACCCCTATGGTCACAGAGAGGGGCTGGAAAAGTATTCGGTAGAGGAAAAAACCGGAGACGTCCTATGCCAAGTCTCTCCGAATGTGGCGAAAGTGACGAAGTGGACCAAGACGAGGCTGCCGGCGATTACGTGTTCCGTATTGTCAACAACGCTATAGCCGACAGAGAAGCAATGCGTCATTCTCTTCTAGAACGAGCAGTTCAGTCGCCTCCGCGCACTCCACGCCGGCCTCTAGCATCAACGTCTACTACTTCCTCTGATTCGTCAACAATGTCAGTCGATTGTCCACCTTCGGCTGGATTTAGTGGGCTTCCTCCGATAACACCAGCTCCACACCAGCCTGTTGAACAGACTGCGTCCATTGAACTGGTATGCAGTACGATGCGTAGACAGATAATATCGAGGGCGTTCTACGGTTGGCTCGCGTATTGCCGACACTTGTCCACTGTCAGAACCCATCTAAGCGGTCTCGTGCATCCCAATATAATTACGAAAGAAGGAGCAGACTCTGGTCTTACAGAAGAAATATGGCGTTCAATGACCGACGACAGTGGTACTGTAAATGACAAAAACGAAGTATACAGGCTGGTTTACTACGGTGGTGTTCAGCATGACATCAGAAAGGAAGTTTGGCCATATCTCCTTGGATATTATGAATTCGGTTCAACAACAGAACAAAGAATAGAACAGGATGCGTCTTGCCGTCGTCAGTACGAGACCACAATGTCGGAGTGGCTCTGTGTTGAAGCCATAGTGCGGCAGAAGGACAGAGAAGCCACGGCCGCTTCAATAGCGAGGCTCAGTGAAGCCTTGGGAAAACTGCCACCCGTGGAGAGTAATGGAGATTCTGGAGAGGTTTTCGAAGACGATTGCAGCGTCATATCAGACAACCCTCCAATTGTAACTCCAGAGCCTAGCGATAACGAACAAAGACGACCGGACAACAAACCTGTTCTGTCTCGAGCACCCAGCGTAGATGAGATTGACAATATCGATTTAGACTCGGAAGAGAAAGAGAAAGATGTTAAAGTTAACGGAGAGACTGACACGAGTGATACTAGAGAAATAGATAGTCTTAAAGACTTAGATGATGAAGATGACGATGTTCATTTAAAAAGCGTTGTGGAGAACCCGGATATGAGGCGAGAAAGTATAGCTGAAATAAAAAGACTGGCTGAAGGTATCGTTCAGAAAAGCGACGGAAGAGGATTACTTTGTAGCGTTGATAGTGCGAATGTGAACCTTAACGGCGTTGACCAAAGATCCTCTGTAGAGGAAAACGGTCCGAGTCCACAACAGCATACTAGCGTTATAGTTACTAATCCGTCTGTTGATCTAGTACCGTCGGGATCACCGGCTTCCGGTGGAACAACTGCTAATGTGAGCCCGTCTCGTAGTCCATTAGGAGTGGTACGAGAAGAATCTCAATCTGCTGGTGCATCATTTGACACTCTGGAACCAACGAGTGATCAACGTCCAGAGACCAGATCCAATTGCGTTTCACCTGCTAGTTCTAATGGTGGAGTTTATTCTAACGAGTTGGTTGAAAGCTTCGCTCTGAATCTCCATCGGATCGAGAAAGACGTTCAAAGATGTGACAGGAATTATCCGTTCTTTACTGATGAAAATCTTGATAAACTAAGAAATATTATGTGCACATACGTTTGGGAACACCTCGAAACTGGTTATATGCAGGGAATGTGTGACCTCGCTGCTCCGCTGCTGGTGATGATGAAGGAGGAAGCTTCAGCCCACGCACTGTTCACAAGGCTCATGGACCGAGCCAGGGATAATTTCCCTTCAGGGCAAGCCATGGACGCTCATTTTGCTGACATGAGGTCGTTAATCCAAATCTTGGACTGCGAGCTGTACGAACTGATGCACGCTCACGGGGATTACACACACTTCTACTTCTGCTATCGCTGGTTCCTATTAGACTTCAAAAGGGAAATGCTATATCAAGACGTGTTTGCGGCGTGGGAAATGATATGGGCTGCTCGTCACGTCGCCTCCGAGCATATGGTGCTGTTCATAGCGCTCGCTCTGCTCGAGACGTATCGCGATGTGATACTCGCGAACACAATGGATTTTACGGATATCATCAAATTCTTCAATGAGATGGCCGAACGTCACGACGCTTCCGCTGTTCTATCTCTAGCCAGAGACCTAGTCCTGCAAGTTCAGACTCTAATCGAGAACAAATAG
Protein
MPAANAPTDPETEQKERLIAAVKKEVKQLMEEAVTRKFVHEDSGGVTTLCGAVEACLGQGLRRRALGLFKTSSTTALLHKIAKHCPEAALVSARVLAAEGTPATRSASGVERRPSAPRPPLCKRGSGSLLAQSPPQSKYLWIRIALFEKQLAKIIEHLVTNATRYYERDALVADPDYGSILSSLLVGPCALEYSKAKPPVTFWTDPPADELVQRHRMSAGTTTPPSVRRPILNFKRSLNASSDDSSSVNTVTGKGTTSAKDYVESLHQNSTATLLYGKNNVRVQPKDAEEPMAGYLSLHQTAVGLVIKWTPNQLMNGYAESESVDKSVYWAMSLQVQVWEVVYVHVHRSQGSDALILVGQDGVQRPPIHFPKGGHLLSFLSNLETGLLPHGQLDPPLWSQRGAGKVFGRGKNRRRPMPSLSECGESDEVDQDEAAGDYVFRIVNNAIADREAMRHSLLERAVQSPPRTPRRPLASTSTTSSDSSTMSVDCPPSAGFSGLPPITPAPHQPVEQTASIELVCSTMRRQIISRAFYGWLAYCRHLSTVRTHLSGLVHPNIITKEGADSGLTEEIWRSMTDDSGTVNDKNEVYRLVYYGGVQHDIRKEVWPYLLGYYEFGSTTEQRIEQDASCRRQYETTMSEWLCVEAIVRQKDREATAASIARLSEALGKLPPVESNGDSGEVFEDDCSVISDNPPIVTPEPSDNEQRRPDNKPVLSRAPSVDEIDNIDLDSEEKEKDVKVNGETDTSDTREIDSLKDLDDEDDDVHLKSVVENPDMRRESIAEIKRLAEGIVQKSDGRGLLCSVDSANVNLNGVDQRSSVEENGPSPQQHTSVIVTNPSVDLVPSGSPASGGTTANVSPSRSPLGVVREESQSAGASFDTLEPTSDQRPETRSNCVSPASSNGGVYSNELVESFALNLHRIEKDVQRCDRNYPFFTDENLDKLRNIMCTYVWEHLETGYMQGMCDLAAPLLVMMKEEASAHALFTRLMDRARDNFPSGQAMDAHFADMRSLIQILDCELYELMHAHGDYTHFYFCYRWFLLDFKREMLYQDVFAAWEMIWAARHVASEHMVLFIALALLETYRDVILANTMDFTDIIKFFNEMAERHDASAVLSLARDLVLQVQTLIENK
Summary
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Cofactor
Mg(2+)
Similarity
Belongs to the helicase family.
Uniprot
H9JTV9
A0A437BAS2
A0A194Q005
A0A212FMJ1
A0A194R1Y5
A0A2A4K274
+ More
A0A2A4K155 A0A2H1VIM7 A0A2H1VE06 A0A084VAK3 A0A182J5U5 A0A1S4N3C9 A0A182MPI1 E0W3Q7 A0A336LLF9 W8B011 X1WJA9 Q8IQ30 E1NZD0 A0A2S2Q3D9 A0A2H8TFH8 A0A2T7PE07 A0A341B397 A0A341B548 A0A091ER45 A0A093NR01 A0A1U7V1Z2 A0A091MS11 A0A3L8SBN6 K7GCK8 K7GCL5 A0A1S3WE58 A0A3M0JY23 A0A3B3CYA9 U3K1V7 A0A094K648 A0A3Q4B3Z4 A0A3B1KB40 A0A341B371 A0A3Q3ETD1 H0ZAZ2 A0A0P5TN43 A0A0P5X0B9 A0A226E6I6 A0A1D2NBK5
A0A2A4K155 A0A2H1VIM7 A0A2H1VE06 A0A084VAK3 A0A182J5U5 A0A1S4N3C9 A0A182MPI1 E0W3Q7 A0A336LLF9 W8B011 X1WJA9 Q8IQ30 E1NZD0 A0A2S2Q3D9 A0A2H8TFH8 A0A2T7PE07 A0A341B397 A0A341B548 A0A091ER45 A0A093NR01 A0A1U7V1Z2 A0A091MS11 A0A3L8SBN6 K7GCK8 K7GCL5 A0A1S3WE58 A0A3M0JY23 A0A3B3CYA9 U3K1V7 A0A094K648 A0A3Q4B3Z4 A0A3B1KB40 A0A341B371 A0A3Q3ETD1 H0ZAZ2 A0A0P5TN43 A0A0P5X0B9 A0A226E6I6 A0A1D2NBK5
EC Number
3.6.4.12
Pubmed
EMBL
BABH01003670
RSAL01000102
RVE47455.1
KQ459582
KPI98912.1
AGBW02007651
+ More
OWR54946.1 KQ460878 KPJ11534.1 NWSH01000252 PCG78004.1 PCG78005.1 ODYU01002776 SOQ40689.1 ODYU01002039 SOQ39047.1 ATLV01004128 KE524190 KFB34997.1 AAZO01007430 AXCM01007464 DS235882 EEB20263.1 UFQS01000045 UFQT01000045 SSW98424.1 SSX18810.1 GAMC01014693 JAB91862.1 ABLF02028172 ABLF02028173 ABLF02028174 ABLF02028176 ABLF02028178 ABLF02030878 ABLF02046579 AE014298 AAN09550.3 AHN59964.1 BT125679 ADN95587.1 GGMS01002519 MBY71722.1 GFXV01001024 MBW12829.1 PZQS01000004 PVD31649.1 KK718897 KFO59486.1 KL224689 KFW62572.1 KK836424 KFP80141.1 QUSF01000030 RLV99772.1 AGCU01134446 AGCU01134447 AGCU01134448 AGCU01134449 AGCU01134450 AGCU01134451 AGCU01134452 QRBI01000121 RMC05843.1 AGTO01020432 KL342336 KFZ53663.1 ABQF01029004 ABQF01029005 ABQF01029006 ABQF01029007 GDIP01124366 JAL79348.1 GDIP01079681 JAM24034.1 LNIX01000006 OXA52958.1 LJIJ01000103 ODN02621.1
OWR54946.1 KQ460878 KPJ11534.1 NWSH01000252 PCG78004.1 PCG78005.1 ODYU01002776 SOQ40689.1 ODYU01002039 SOQ39047.1 ATLV01004128 KE524190 KFB34997.1 AAZO01007430 AXCM01007464 DS235882 EEB20263.1 UFQS01000045 UFQT01000045 SSW98424.1 SSX18810.1 GAMC01014693 JAB91862.1 ABLF02028172 ABLF02028173 ABLF02028174 ABLF02028176 ABLF02028178 ABLF02030878 ABLF02046579 AE014298 AAN09550.3 AHN59964.1 BT125679 ADN95587.1 GGMS01002519 MBY71722.1 GFXV01001024 MBW12829.1 PZQS01000004 PVD31649.1 KK718897 KFO59486.1 KL224689 KFW62572.1 KK836424 KFP80141.1 QUSF01000030 RLV99772.1 AGCU01134446 AGCU01134447 AGCU01134448 AGCU01134449 AGCU01134450 AGCU01134451 AGCU01134452 QRBI01000121 RMC05843.1 AGTO01020432 KL342336 KFZ53663.1 ABQF01029004 ABQF01029005 ABQF01029006 ABQF01029007 GDIP01124366 JAL79348.1 GDIP01079681 JAM24034.1 LNIX01000006 OXA52958.1 LJIJ01000103 ODN02621.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000007151
UP000053240
UP000218220
+ More
UP000030765 UP000075880 UP000075883 UP000009046 UP000007819 UP000000803 UP000245119 UP000252040 UP000052976 UP000054081 UP000189704 UP000276834 UP000007267 UP000079721 UP000269221 UP000261560 UP000016665 UP000261620 UP000018467 UP000261660 UP000007754 UP000198287 UP000094527
UP000030765 UP000075880 UP000075883 UP000009046 UP000007819 UP000000803 UP000245119 UP000252040 UP000052976 UP000054081 UP000189704 UP000276834 UP000007267 UP000079721 UP000269221 UP000261560 UP000016665 UP000261620 UP000018467 UP000261660 UP000007754 UP000198287 UP000094527
Interpro
CDD
ProteinModelPortal
H9JTV9
A0A437BAS2
A0A194Q005
A0A212FMJ1
A0A194R1Y5
A0A2A4K274
+ More
A0A2A4K155 A0A2H1VIM7 A0A2H1VE06 A0A084VAK3 A0A182J5U5 A0A1S4N3C9 A0A182MPI1 E0W3Q7 A0A336LLF9 W8B011 X1WJA9 Q8IQ30 E1NZD0 A0A2S2Q3D9 A0A2H8TFH8 A0A2T7PE07 A0A341B397 A0A341B548 A0A091ER45 A0A093NR01 A0A1U7V1Z2 A0A091MS11 A0A3L8SBN6 K7GCK8 K7GCL5 A0A1S3WE58 A0A3M0JY23 A0A3B3CYA9 U3K1V7 A0A094K648 A0A3Q4B3Z4 A0A3B1KB40 A0A341B371 A0A3Q3ETD1 H0ZAZ2 A0A0P5TN43 A0A0P5X0B9 A0A226E6I6 A0A1D2NBK5
A0A2A4K155 A0A2H1VIM7 A0A2H1VE06 A0A084VAK3 A0A182J5U5 A0A1S4N3C9 A0A182MPI1 E0W3Q7 A0A336LLF9 W8B011 X1WJA9 Q8IQ30 E1NZD0 A0A2S2Q3D9 A0A2H8TFH8 A0A2T7PE07 A0A341B397 A0A341B548 A0A091ER45 A0A093NR01 A0A1U7V1Z2 A0A091MS11 A0A3L8SBN6 K7GCK8 K7GCL5 A0A1S3WE58 A0A3M0JY23 A0A3B3CYA9 U3K1V7 A0A094K648 A0A3Q4B3Z4 A0A3B1KB40 A0A341B371 A0A3Q3ETD1 H0ZAZ2 A0A0P5TN43 A0A0P5X0B9 A0A226E6I6 A0A1D2NBK5
PDB
4QXA
E-value=6.77819e-46,
Score=468
Ontologies
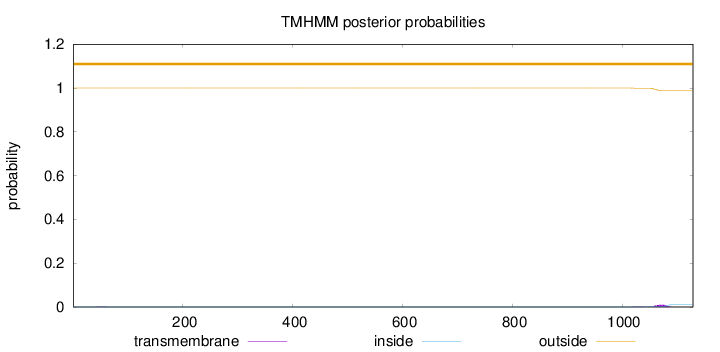
Topology
Length:
1129
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.23775
Exp number, first 60 AAs:
0.00375
Total prob of N-in:
0.00023
outside
1 - 1129
Population Genetic Test Statistics
Pi
183.84666
Theta
167.138829
Tajima's D
0.41315
CLR
0.333005
CSRT
0.489275536223189
Interpretation
Uncertain
