Gene
KWMTBOMO09650
Pre Gene Modal
BGIBMGA012865
Annotation
peptidoglycan_recognition_protein_precursor_[Bombyx_mori]
Full name
Peptidoglycan-recognition protein
Location in the cell
Extracellular Reliability : 1.191 PlasmaMembrane Reliability : 1.444
Sequence
CDS
ATGGCTGGTCTATTTTTATTCATTGCCATCAGCTTAATCTCGGTAGTGACTGCATTACCTAGTCCAATACAAACAGCTAGTGCGTATTCCTTCCCTTTCGTAAGCCGAGAGGAGTGGGGTGCAAGGCCATCGACAGCGTCATCACCCCTTGGAGTTAGTCCAGTGCCCATAGTAGTGATCCATCACAGCTATATTCCCAGAATATGCTTAACTCGGGCAAACTGTGAGAGAGATATGCGCAACATGCAGAGAGTGCACCAAGTAACCAATGGCTGGACAGATATCGGATATCATTTTGCTGTCGGCGGTGAAGGAACTGTCTTCGAGGGCCGTGGATGGAGTACACTTGGAGCTCACACATACGGCGTCAACACCCGAAGCATCGGAATCTTGTTGATTGGAGATTTTATTACTAATAAACCGCCACAGGCGCAGCTACAATCCGTCAAGGACCTGATCGAAGCTGGTGTGAGGCTCGGTCACATCCGATCTGATTACAAACTCGTCGGACACAGACAAGTCACTCCCACTGAATGCCCTGGACAGAGATTGTTTGATGAAATCTCAAAATGGGATCACTTTTCACTTAATTGGGATTGA
Protein
MAGLFLFIAISLISVVTALPSPIQTASAYSFPFVSREEWGARPSTASSPLGVSPVPIVVIHHSYIPRICLTRANCERDMRNMQRVHQVTNGWTDIGYHFAVGGEGTVFEGRGWSTLGAHTYGVNTRSIGILLIGDFITNKPPQAQLQSVKDLIEAGVRLGHIRSDYKLVGHRQVTPTECPGQRLFDEISKWDHFSLNWD
Summary
Similarity
Belongs to the N-acetylmuramoyl-L-alanine amidase 2 family.
Feature
chain Peptidoglycan-recognition protein
Uniprot
G8GBC2
Q0KKW7
D1KRK9
I7FE39
A0A125S9Y3
A0A0K8TH54
+ More
A0A2A4K1S1 A0A2S1GUY3 A0A2H1VA87 E7DN57 A0A194R6P0 A0A194Q1P4 A0A0N0PA57 A0A194R2Q4 A0A194Q679 A0A2A4K1V5 A0A437BAM8 A0A346RAD1 W8R8R8 A0A194R163 A0A212FMH5 A7BIV1 A0A0N1PHT4 A0A194PRQ4 E7DN59 A0A0N1I4J1 A0A2H1VU37 A0A437BAT0 D2A6F2 A0A346RAC8 A0A3S2L7J3 E7DN58 A0A1E1W0W1 A0A212FMI9 A0A194Q0S5 T1PIP5 A0A1I8NHG6 A0A2A4K187 A0A034VAP3 A0A1E1WD69 I7EUB7 T1E303 A0A1J1HM36 A0A0K8UF23 A0A0K8VL70 A0A1Y1JSG0 M1FW43 A0A1Y1LEP6 B4NAW7 A0A1Y1LES5 A0A0Q9X525 A0A1Q3FPI9 A0A1Q3FPS8 B6RQQ1 A0A0A1WUF7 A0A411IZR2 A0A411J001 Q9BLL2 A0A411IZU6 A0A0K0TN30 A0A1L8E3Q3 A0A1J0M178 A0A0A1WR50 A0A0L0BUT1 W8C339 E5D601 W8BGX0 Q0KKW6 A0A023ELI4 A0A1S4FPG4 A0A023EI58 Q1HRH3 A0A1Q3FR98 U3Q0G7 A0A0Q9XDH5 A0A0M4EHG6 B4KDZ3 A0A1W7R8P4 A0A182GJG8 A0A1I8QBA4 A8D1K9 A0A2S1UFA9 A0A1W4W1D6 A0A2M3YYC2 A0A1W4XLB6 A0A1W4WC29 A0A1W4WDD2 A0A1W4XVT8 U4UXK7 A0A1W4XWP1 A0A182VW14 A0A182YHW5 A0A1E1WV23 F4MI27 B3NZE4 B3M0X2 J3JTK0 B4JUP2 A0A182SIN9 A0A3F2YX68
A0A2A4K1S1 A0A2S1GUY3 A0A2H1VA87 E7DN57 A0A194R6P0 A0A194Q1P4 A0A0N0PA57 A0A194R2Q4 A0A194Q679 A0A2A4K1V5 A0A437BAM8 A0A346RAD1 W8R8R8 A0A194R163 A0A212FMH5 A7BIV1 A0A0N1PHT4 A0A194PRQ4 E7DN59 A0A0N1I4J1 A0A2H1VU37 A0A437BAT0 D2A6F2 A0A346RAC8 A0A3S2L7J3 E7DN58 A0A1E1W0W1 A0A212FMI9 A0A194Q0S5 T1PIP5 A0A1I8NHG6 A0A2A4K187 A0A034VAP3 A0A1E1WD69 I7EUB7 T1E303 A0A1J1HM36 A0A0K8UF23 A0A0K8VL70 A0A1Y1JSG0 M1FW43 A0A1Y1LEP6 B4NAW7 A0A1Y1LES5 A0A0Q9X525 A0A1Q3FPI9 A0A1Q3FPS8 B6RQQ1 A0A0A1WUF7 A0A411IZR2 A0A411J001 Q9BLL2 A0A411IZU6 A0A0K0TN30 A0A1L8E3Q3 A0A1J0M178 A0A0A1WR50 A0A0L0BUT1 W8C339 E5D601 W8BGX0 Q0KKW6 A0A023ELI4 A0A1S4FPG4 A0A023EI58 Q1HRH3 A0A1Q3FR98 U3Q0G7 A0A0Q9XDH5 A0A0M4EHG6 B4KDZ3 A0A1W7R8P4 A0A182GJG8 A0A1I8QBA4 A8D1K9 A0A2S1UFA9 A0A1W4W1D6 A0A2M3YYC2 A0A1W4XLB6 A0A1W4WC29 A0A1W4WDD2 A0A1W4XVT8 U4UXK7 A0A1W4XWP1 A0A182VW14 A0A182YHW5 A0A1E1WV23 F4MI27 B3NZE4 B3M0X2 J3JTK0 B4JUP2 A0A182SIN9 A0A3F2YX68
Pubmed
EMBL
JN120254
AER70118.1
AB232693
BAF03520.1
GQ293365
ACX49764.1
+ More
JX082166 AFP23116.1 KT725232 AME17979.1 GBRD01001336 JAG64485.1 NWSH01000252 PCG78009.1 MH078491 AWE05152.1 ODYU01001490 SOQ37763.1 HQ260964 ADU33185.1 KQ460878 KPJ11531.1 KQ459582 KPI98914.1 KQ458843 KPJ04889.1 KPJ11530.1 KPI98915.1 PCG78006.1 RSAL01000102 RVE47454.1 MH200921 AXS59131.1 KF985962 AHL58837.1 KPJ11533.1 AGBW02007651 OWR54948.1 AB291943 BAF74637.1 KQ458412 KPJ05906.1 KQ459595 KPI95992.1 HQ260966 ADU33187.1 KPJ05907.1 ODYU01004458 SOQ44347.1 RVE47453.1 KQ971347 EFA05744.1 MH200918 AXS59128.1 RVE47452.1 HQ260965 ADU33186.1 GDQN01010451 JAT80603.1 OWR54947.1 KPI98913.1 KA647970 AFP62599.1 PCG78007.1 GAKP01019368 GAKP01019367 JAC39584.1 GDQN01006138 JAT84916.1 JX082167 AFP23117.1 GALA01000820 JAA94032.1 CVRI01000010 CRK88995.1 GDHF01027116 JAI25198.1 GDHF01033343 GDHF01012683 JAI18971.1 JAI39631.1 GEZM01102141 JAV51943.1 JQ687224 AFV15800.1 GEZM01057719 JAV72122.1 CH964232 EDW80931.1 GEZM01057718 JAV72123.1 KRF99362.1 GFDL01005652 JAV29393.1 GFDL01005478 JAV29567.1 EU282122 ABZ80672.1 GBXI01011810 JAD02482.1 MH423627 QBC16601.1 MH423626 QBC16600.1 AB017519 BAB33294.1 MH423628 QBC16602.1 KR534884 AKR52932.1 GFDF01000959 JAV13125.1 KX342009 APD13821.1 GBXI01013394 JAD00898.1 JRES01001304 KNC23748.1 GAMC01010331 JAB96224.1 GU182906 ADR51144.1 GAMC01010332 GAMC01010330 GAMC01010329 JAB96223.1 AB232694 BAF03521.1 GAPW01004489 JAC09109.1 GAPW01004525 JAC09073.1 DQ440121 CH477643 ABF18154.1 EAT37875.1 GFDL01005049 JAV29996.1 KF282764 AGW80663.1 CH933806 KRG01864.1 CP012526 ALC45808.1 EDW16014.1 GEHC01000124 JAV47521.1 JXUM01068194 KQ562487 KXJ75805.1 AJVK01004217 EU130784 ABV60369.1 MG457266 AWI97805.1 GGFM01000499 MBW21250.1 KB632404 ERL95020.1 GDQN01000222 JAT90832.1 EZ617708 ADH94601.1 CH954181 EDV49517.1 CH902617 EDV43201.1 BT126557 AEE61521.1 CH916374 EDV91212.1 AXCN02001500
JX082166 AFP23116.1 KT725232 AME17979.1 GBRD01001336 JAG64485.1 NWSH01000252 PCG78009.1 MH078491 AWE05152.1 ODYU01001490 SOQ37763.1 HQ260964 ADU33185.1 KQ460878 KPJ11531.1 KQ459582 KPI98914.1 KQ458843 KPJ04889.1 KPJ11530.1 KPI98915.1 PCG78006.1 RSAL01000102 RVE47454.1 MH200921 AXS59131.1 KF985962 AHL58837.1 KPJ11533.1 AGBW02007651 OWR54948.1 AB291943 BAF74637.1 KQ458412 KPJ05906.1 KQ459595 KPI95992.1 HQ260966 ADU33187.1 KPJ05907.1 ODYU01004458 SOQ44347.1 RVE47453.1 KQ971347 EFA05744.1 MH200918 AXS59128.1 RVE47452.1 HQ260965 ADU33186.1 GDQN01010451 JAT80603.1 OWR54947.1 KPI98913.1 KA647970 AFP62599.1 PCG78007.1 GAKP01019368 GAKP01019367 JAC39584.1 GDQN01006138 JAT84916.1 JX082167 AFP23117.1 GALA01000820 JAA94032.1 CVRI01000010 CRK88995.1 GDHF01027116 JAI25198.1 GDHF01033343 GDHF01012683 JAI18971.1 JAI39631.1 GEZM01102141 JAV51943.1 JQ687224 AFV15800.1 GEZM01057719 JAV72122.1 CH964232 EDW80931.1 GEZM01057718 JAV72123.1 KRF99362.1 GFDL01005652 JAV29393.1 GFDL01005478 JAV29567.1 EU282122 ABZ80672.1 GBXI01011810 JAD02482.1 MH423627 QBC16601.1 MH423626 QBC16600.1 AB017519 BAB33294.1 MH423628 QBC16602.1 KR534884 AKR52932.1 GFDF01000959 JAV13125.1 KX342009 APD13821.1 GBXI01013394 JAD00898.1 JRES01001304 KNC23748.1 GAMC01010331 JAB96224.1 GU182906 ADR51144.1 GAMC01010332 GAMC01010330 GAMC01010329 JAB96223.1 AB232694 BAF03521.1 GAPW01004489 JAC09109.1 GAPW01004525 JAC09073.1 DQ440121 CH477643 ABF18154.1 EAT37875.1 GFDL01005049 JAV29996.1 KF282764 AGW80663.1 CH933806 KRG01864.1 CP012526 ALC45808.1 EDW16014.1 GEHC01000124 JAV47521.1 JXUM01068194 KQ562487 KXJ75805.1 AJVK01004217 EU130784 ABV60369.1 MG457266 AWI97805.1 GGFM01000499 MBW21250.1 KB632404 ERL95020.1 GDQN01000222 JAT90832.1 EZ617708 ADH94601.1 CH954181 EDV49517.1 CH902617 EDV43201.1 BT126557 AEE61521.1 CH916374 EDV91212.1 AXCN02001500
Proteomes
UP000218220
UP000053240
UP000053268
UP000283053
UP000007151
UP000007266
+ More
UP000095301 UP000183832 UP000007798 UP000037069 UP000008820 UP000009192 UP000092553 UP000069940 UP000249989 UP000095300 UP000092462 UP000192221 UP000192223 UP000030742 UP000075920 UP000076408 UP000008711 UP000007801 UP000001070 UP000075901 UP000075886
UP000095301 UP000183832 UP000007798 UP000037069 UP000008820 UP000009192 UP000092553 UP000069940 UP000249989 UP000095300 UP000092462 UP000192221 UP000192223 UP000030742 UP000075920 UP000076408 UP000008711 UP000007801 UP000001070 UP000075901 UP000075886
Pfam
PF01510 Amidase_2
Interpro
SUPFAM
SSF55846
SSF55846
Gene 3D
CDD
ProteinModelPortal
G8GBC2
Q0KKW7
D1KRK9
I7FE39
A0A125S9Y3
A0A0K8TH54
+ More
A0A2A4K1S1 A0A2S1GUY3 A0A2H1VA87 E7DN57 A0A194R6P0 A0A194Q1P4 A0A0N0PA57 A0A194R2Q4 A0A194Q679 A0A2A4K1V5 A0A437BAM8 A0A346RAD1 W8R8R8 A0A194R163 A0A212FMH5 A7BIV1 A0A0N1PHT4 A0A194PRQ4 E7DN59 A0A0N1I4J1 A0A2H1VU37 A0A437BAT0 D2A6F2 A0A346RAC8 A0A3S2L7J3 E7DN58 A0A1E1W0W1 A0A212FMI9 A0A194Q0S5 T1PIP5 A0A1I8NHG6 A0A2A4K187 A0A034VAP3 A0A1E1WD69 I7EUB7 T1E303 A0A1J1HM36 A0A0K8UF23 A0A0K8VL70 A0A1Y1JSG0 M1FW43 A0A1Y1LEP6 B4NAW7 A0A1Y1LES5 A0A0Q9X525 A0A1Q3FPI9 A0A1Q3FPS8 B6RQQ1 A0A0A1WUF7 A0A411IZR2 A0A411J001 Q9BLL2 A0A411IZU6 A0A0K0TN30 A0A1L8E3Q3 A0A1J0M178 A0A0A1WR50 A0A0L0BUT1 W8C339 E5D601 W8BGX0 Q0KKW6 A0A023ELI4 A0A1S4FPG4 A0A023EI58 Q1HRH3 A0A1Q3FR98 U3Q0G7 A0A0Q9XDH5 A0A0M4EHG6 B4KDZ3 A0A1W7R8P4 A0A182GJG8 A0A1I8QBA4 A8D1K9 A0A2S1UFA9 A0A1W4W1D6 A0A2M3YYC2 A0A1W4XLB6 A0A1W4WC29 A0A1W4WDD2 A0A1W4XVT8 U4UXK7 A0A1W4XWP1 A0A182VW14 A0A182YHW5 A0A1E1WV23 F4MI27 B3NZE4 B3M0X2 J3JTK0 B4JUP2 A0A182SIN9 A0A3F2YX68
A0A2A4K1S1 A0A2S1GUY3 A0A2H1VA87 E7DN57 A0A194R6P0 A0A194Q1P4 A0A0N0PA57 A0A194R2Q4 A0A194Q679 A0A2A4K1V5 A0A437BAM8 A0A346RAD1 W8R8R8 A0A194R163 A0A212FMH5 A7BIV1 A0A0N1PHT4 A0A194PRQ4 E7DN59 A0A0N1I4J1 A0A2H1VU37 A0A437BAT0 D2A6F2 A0A346RAC8 A0A3S2L7J3 E7DN58 A0A1E1W0W1 A0A212FMI9 A0A194Q0S5 T1PIP5 A0A1I8NHG6 A0A2A4K187 A0A034VAP3 A0A1E1WD69 I7EUB7 T1E303 A0A1J1HM36 A0A0K8UF23 A0A0K8VL70 A0A1Y1JSG0 M1FW43 A0A1Y1LEP6 B4NAW7 A0A1Y1LES5 A0A0Q9X525 A0A1Q3FPI9 A0A1Q3FPS8 B6RQQ1 A0A0A1WUF7 A0A411IZR2 A0A411J001 Q9BLL2 A0A411IZU6 A0A0K0TN30 A0A1L8E3Q3 A0A1J0M178 A0A0A1WR50 A0A0L0BUT1 W8C339 E5D601 W8BGX0 Q0KKW6 A0A023ELI4 A0A1S4FPG4 A0A023EI58 Q1HRH3 A0A1Q3FR98 U3Q0G7 A0A0Q9XDH5 A0A0M4EHG6 B4KDZ3 A0A1W7R8P4 A0A182GJG8 A0A1I8QBA4 A8D1K9 A0A2S1UFA9 A0A1W4W1D6 A0A2M3YYC2 A0A1W4XLB6 A0A1W4WC29 A0A1W4WDD2 A0A1W4XVT8 U4UXK7 A0A1W4XWP1 A0A182VW14 A0A182YHW5 A0A1E1WV23 F4MI27 B3NZE4 B3M0X2 J3JTK0 B4JUP2 A0A182SIN9 A0A3F2YX68
PDB
1OHT
E-value=3.32703e-44,
Score=445
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.986290
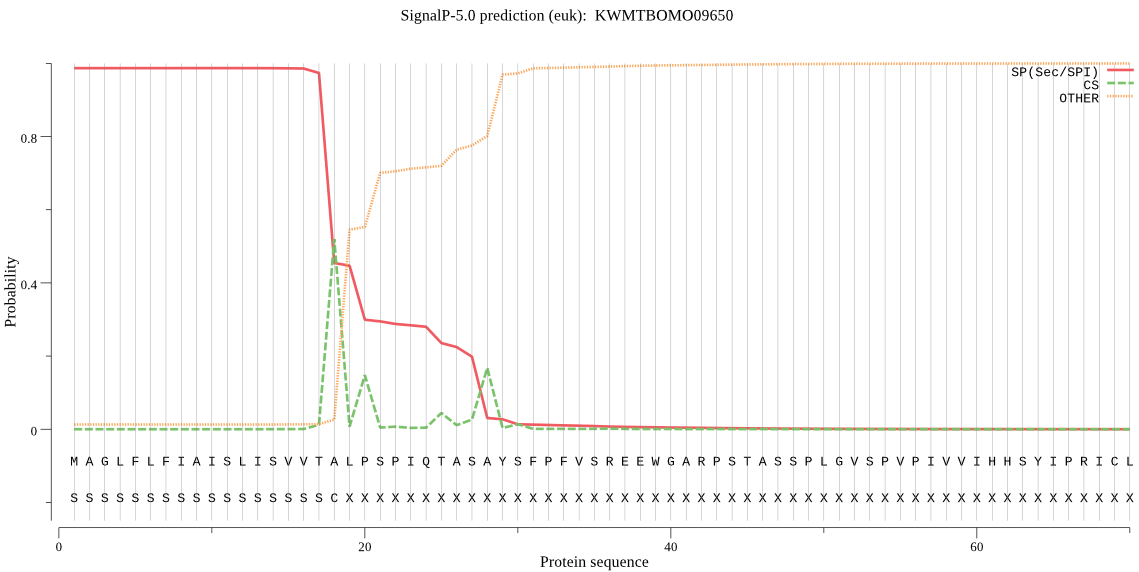
Length:
199
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
17.74692
Exp number, first 60 AAs:
16.67305
Total prob of N-in:
0.65739
POSSIBLE N-term signal
sequence
outside
1 - 199

Population Genetic Test Statistics
Pi
216.038629
Theta
167.601557
Tajima's D
1.020144
CLR
0
CSRT
0.667116644167792
Interpretation
Uncertain
