Gene
KWMTBOMO09641 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012859
Annotation
NIPSNAP_protein_[Bombyx_mori]
Full name
Protein NipSnap
Location in the cell
Mitochondrial Reliability : 2.543
Sequence
CDS
ATGAATTCCCTTAATAAGTTATCAGTTTATAGTAAATTAGTGCCGAAAAATGCCAGGGTGATATCTACAAGTGCACTATTAAACACTGACGATGGATGGTTTTCGAAATTACTAGTTCGAAGAATCGAACCGACAAAAGAGTCACACAGTCGAATGCTTAGTGACAAGGAAGTAATTTATGCTCTTCATACACACAACATAAGACCGGATTCAGTGGACAACTATTTAAAAAATTACAAGCAGCATGTAGATCTAATACATTCACACAAAGCAGAACTTGGTTGTGAGCTTGTAGGATCATGGACAGTATCAGTTGGAGATATGGATCAAGCTCTGCATTTATTCAAATATGTTGGTGGATTTGAAAAAATTGACAAGGCCAAACAATTGTTTAAACAAGATTCTAGTTACCAATTGCTAGAAAGTGAGAGAGGCAAAATGGTCAGGTCCAGACATCTCCAATACTTGCTAGCTTTCAGCTTTTGGCCAGCTGGAGAAGTACGGTCTCCCAACAACATCTACGAAATACGATCATACAGTTTAAAACCTGGAACTATGATTGAGTGGGGTAACAATTGGGCGCGTGGCCTTACTTACCGACGCGCGCAAAACGAGGCGTTTGCGGGCTATTTCTCACAAATTGGCCGATTATACAATGTTCATCATATTTGGTGCTATAAAGATTTGCAAGCTCGTCGTGAAACTAGAGAGAGTACATGGCGTAATCCAGGCTGGGATGAGTGTGTCGCGTACACGGTGCCTCTGATCAGAGAGATGCACTGTCGTATCTTAGAACCCACTGAATTCTCTCCTACACAATAA
Protein
MNSLNKLSVYSKLVPKNARVISTSALLNTDDGWFSKLLVRRIEPTKESHSRMLSDKEVIYALHTHNIRPDSVDNYLKNYKQHVDLIHSHKAELGCELVGSWTVSVGDMDQALHLFKYVGGFEKIDKAKQLFKQDSSYQLLESERGKMVRSRHLQYLLAFSFWPAGEVRSPNNIYEIRSYSLKPGTMIEWGNNWARGLTYRRAQNEAFAGYFSQIGRLYNVHHIWCYKDLQARRETRESTWRNPGWDECVAYTVPLIREMHCRILEPTEFSPTQ
Summary
Similarity
Belongs to the NipSnap family.
Keywords
Complete proteome
Reference proteome
Feature
chain Protein NipSnap
Uniprot
Q2F671
A0A2H1VIT7
A0A2A4JK64
A0A194Q1Q4
A0A194RSL7
A0A212FMJ6
+ More
A0A437BIN3 S4PD61 A0A0L7L306 A0A0J7NWY4 E2ANA8 E9IVX2 A0A026WEC4 A0A151WY94 F4WQ54 A0A151I5Y0 A0A158NX73 A0A195EDJ3 B4NET2 A0A1I8MNN4 A0A1W4UN95 A0A151JU04 B4JLK2 A0A232FBS8 K7J4E3 A0A1W4V295 B4R5Q9 U5EIJ3 A0A1L8EE23 A0A0C9RD66 B4MGB9 M9PJP0 Q9VXK0 A0A151I798 B4IL16 A0A1L8EAD3 A0A0Q9XSZ8 B4L7M8 A0A0Q9XG84 A0A3B0K6P1 B4H134 A0A182M5F8 B3NXF3 Q29GK0 A0A1B6CJG4 D3TMS9 A0A1A9ZLF5 A0A1I8Q4E5 W8BXN9 A0A1B0C564 A0A0A1WUC4 A0A1A9W645 B4PXB5 A0A0K8VWH4 A0A182RC46 A0A034WIA3 A0A0C9R3Y7 A0A2M3Z567 A0A182YHI0 A0A182V8Z3 A0A182XE39 Q7PGV6 A0A067QIR2 A0A2M3Z573 A0A182SCL7 A0A2M4AGT3 A0A0K8TP78 A0A182JWB1 A0A1B6F0A0 A0A2M4BWM2 A0A2M4AGY6 A0A2M4BWF4 A0A1L8DW63 A0A182JIL2 T1DNR2 A0A182PT26 W5JDE4 A0A182FKU5 A0A182PZI3 A0A1B6MD62 A0A182LCF0 A0A182HJ84 A0A084VAK1 A0A182N509 A0A182UBT9 A0A023EM56 A0A2M4ALA4 A0A1Q3FHB1 Q17Q08 A0A1S4EV42 A0A1L8DW49 B0X2Y6 A0A336LQI5 A0A023F895 R4FM21 R4WD15 A0A069DRZ0 E0VR64 A0A1Y1KH08 A0A1W4X9P5
A0A437BIN3 S4PD61 A0A0L7L306 A0A0J7NWY4 E2ANA8 E9IVX2 A0A026WEC4 A0A151WY94 F4WQ54 A0A151I5Y0 A0A158NX73 A0A195EDJ3 B4NET2 A0A1I8MNN4 A0A1W4UN95 A0A151JU04 B4JLK2 A0A232FBS8 K7J4E3 A0A1W4V295 B4R5Q9 U5EIJ3 A0A1L8EE23 A0A0C9RD66 B4MGB9 M9PJP0 Q9VXK0 A0A151I798 B4IL16 A0A1L8EAD3 A0A0Q9XSZ8 B4L7M8 A0A0Q9XG84 A0A3B0K6P1 B4H134 A0A182M5F8 B3NXF3 Q29GK0 A0A1B6CJG4 D3TMS9 A0A1A9ZLF5 A0A1I8Q4E5 W8BXN9 A0A1B0C564 A0A0A1WUC4 A0A1A9W645 B4PXB5 A0A0K8VWH4 A0A182RC46 A0A034WIA3 A0A0C9R3Y7 A0A2M3Z567 A0A182YHI0 A0A182V8Z3 A0A182XE39 Q7PGV6 A0A067QIR2 A0A2M3Z573 A0A182SCL7 A0A2M4AGT3 A0A0K8TP78 A0A182JWB1 A0A1B6F0A0 A0A2M4BWM2 A0A2M4AGY6 A0A2M4BWF4 A0A1L8DW63 A0A182JIL2 T1DNR2 A0A182PT26 W5JDE4 A0A182FKU5 A0A182PZI3 A0A1B6MD62 A0A182LCF0 A0A182HJ84 A0A084VAK1 A0A182N509 A0A182UBT9 A0A023EM56 A0A2M4ALA4 A0A1Q3FHB1 Q17Q08 A0A1S4EV42 A0A1L8DW49 B0X2Y6 A0A336LQI5 A0A023F895 R4FM21 R4WD15 A0A069DRZ0 E0VR64 A0A1Y1KH08 A0A1W4X9P5
Pubmed
26354079
22118469
23622113
26227816
20798317
21282665
+ More
24508170 30249741 21719571 21347285 17994087 25315136 28648823 20075255 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 15632085 23185243 20353571 24495485 25830018 17550304 25348373 25244985 12364791 14747013 17210077 24845553 26369729 20920257 23761445 20966253 24438588 24945155 26483478 17510324 25474469 23691247 26334808 20566863 28004739
24508170 30249741 21719571 21347285 17994087 25315136 28648823 20075255 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 15632085 23185243 20353571 24495485 25830018 17550304 25348373 25244985 12364791 14747013 17210077 24845553 26369729 20920257 23761445 20966253 24438588 24945155 26483478 17510324 25474469 23691247 26334808 20566863 28004739
EMBL
DQ311201
ABD36146.1
ODYU01002803
SOQ40750.1
NWSH01001194
PCG72166.1
+ More
KQ459582 KPI98924.1 KQ459700 KPJ20340.1 AGBW02007651 OWR54957.1 RSAL01000051 RVE50304.1 GAIX01005082 JAA87478.1 JTDY01003286 KOB69800.1 LBMM01001097 KMQ96930.1 GL441177 EFN65056.1 GL766449 EFZ15215.1 KK107260 QOIP01000001 EZA54016.1 RLU27073.1 KQ982651 KYQ52858.1 GL888262 EGI63677.1 KQ976434 KYM87223.1 ADTU01003011 KQ979074 KYN22882.1 CH964239 EDW82251.1 KQ981799 KYN35666.1 CH916370 EDW00455.1 NNAY01000455 OXU28281.1 CM000366 EDX18059.1 GANO01002615 JAB57256.1 GFDG01001829 JAV16970.1 GBYB01006220 JAG75987.1 CH940672 EDW57442.1 KRF77770.1 AE014298 AHN59786.1 AY061048 AJ249798 KQ978464 KYM93772.1 CH480862 EDW52823.1 GFDG01003227 JAV15572.1 CH933814 KRG07511.1 EDW05741.1 KRG07512.1 KRG07510.1 OUUW01000038 SPP89887.1 CH479201 EDW29950.1 AXCM01004774 CH954180 EDV46912.1 KQS30243.1 CH379064 EAL32109.1 KRT06915.1 GEDC01030241 GEDC01023756 GEDC01019150 GEDC01004970 GEDC01001475 JAS07057.1 JAS13542.1 JAS18148.1 JAS32328.1 JAS35823.1 EZ422731 ADD19007.1 GAMC01002478 JAC04078.1 JXJN01025869 GBXI01012172 JAD02120.1 CM000162 EDX01878.1 KRK06410.1 GDHF01009091 GDHF01005223 JAI43223.1 JAI47091.1 GAKP01004885 JAC54067.1 GBYB01007517 JAG77284.1 GGFM01002894 MBW23645.1 AAAB01008879 EAA44790.3 KK853319 KDR08564.1 GGFM01002911 MBW23662.1 GGFK01006670 MBW39991.1 GDAI01001640 JAI15963.1 GECZ01026476 JAS43293.1 GGFJ01008190 MBW57331.1 GGFK01006725 MBW40046.1 GGFJ01008191 MBW57332.1 GFDF01003403 JAV10681.1 GAMD01002640 JAA98950.1 ADMH02001488 ETN62372.1 AXCN02000180 GEBQ01006158 JAT33819.1 APCN01002131 ATLV01004128 KE524190 KFB34995.1 JXUM01110641 GAPW01003227 KQ565430 JAC10371.1 KXJ70984.1 GGFK01008191 MBW41512.1 GFDL01008120 JAV26925.1 CH477188 EAT48786.1 GFDF01003420 JAV10664.1 DS232303 EDS39488.1 UFQS01000045 UFQT01000045 SSW98423.1 SSX18809.1 GBBI01001222 JAC17490.1 ACPB03020346 GAHY01001894 JAA75616.1 AK417318 BAN20533.1 GBGD01002413 JAC86476.1 DS235451 EEB15870.1 GEZM01085934 JAV59878.1
KQ459582 KPI98924.1 KQ459700 KPJ20340.1 AGBW02007651 OWR54957.1 RSAL01000051 RVE50304.1 GAIX01005082 JAA87478.1 JTDY01003286 KOB69800.1 LBMM01001097 KMQ96930.1 GL441177 EFN65056.1 GL766449 EFZ15215.1 KK107260 QOIP01000001 EZA54016.1 RLU27073.1 KQ982651 KYQ52858.1 GL888262 EGI63677.1 KQ976434 KYM87223.1 ADTU01003011 KQ979074 KYN22882.1 CH964239 EDW82251.1 KQ981799 KYN35666.1 CH916370 EDW00455.1 NNAY01000455 OXU28281.1 CM000366 EDX18059.1 GANO01002615 JAB57256.1 GFDG01001829 JAV16970.1 GBYB01006220 JAG75987.1 CH940672 EDW57442.1 KRF77770.1 AE014298 AHN59786.1 AY061048 AJ249798 KQ978464 KYM93772.1 CH480862 EDW52823.1 GFDG01003227 JAV15572.1 CH933814 KRG07511.1 EDW05741.1 KRG07512.1 KRG07510.1 OUUW01000038 SPP89887.1 CH479201 EDW29950.1 AXCM01004774 CH954180 EDV46912.1 KQS30243.1 CH379064 EAL32109.1 KRT06915.1 GEDC01030241 GEDC01023756 GEDC01019150 GEDC01004970 GEDC01001475 JAS07057.1 JAS13542.1 JAS18148.1 JAS32328.1 JAS35823.1 EZ422731 ADD19007.1 GAMC01002478 JAC04078.1 JXJN01025869 GBXI01012172 JAD02120.1 CM000162 EDX01878.1 KRK06410.1 GDHF01009091 GDHF01005223 JAI43223.1 JAI47091.1 GAKP01004885 JAC54067.1 GBYB01007517 JAG77284.1 GGFM01002894 MBW23645.1 AAAB01008879 EAA44790.3 KK853319 KDR08564.1 GGFM01002911 MBW23662.1 GGFK01006670 MBW39991.1 GDAI01001640 JAI15963.1 GECZ01026476 JAS43293.1 GGFJ01008190 MBW57331.1 GGFK01006725 MBW40046.1 GGFJ01008191 MBW57332.1 GFDF01003403 JAV10681.1 GAMD01002640 JAA98950.1 ADMH02001488 ETN62372.1 AXCN02000180 GEBQ01006158 JAT33819.1 APCN01002131 ATLV01004128 KE524190 KFB34995.1 JXUM01110641 GAPW01003227 KQ565430 JAC10371.1 KXJ70984.1 GGFK01008191 MBW41512.1 GFDL01008120 JAV26925.1 CH477188 EAT48786.1 GFDF01003420 JAV10664.1 DS232303 EDS39488.1 UFQS01000045 UFQT01000045 SSW98423.1 SSX18809.1 GBBI01001222 JAC17490.1 ACPB03020346 GAHY01001894 JAA75616.1 AK417318 BAN20533.1 GBGD01002413 JAC86476.1 DS235451 EEB15870.1 GEZM01085934 JAV59878.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000007151
UP000283053
UP000037510
+ More
UP000036403 UP000000311 UP000053097 UP000279307 UP000075809 UP000007755 UP000078540 UP000005205 UP000078492 UP000007798 UP000095301 UP000192221 UP000078541 UP000001070 UP000215335 UP000002358 UP000000304 UP000008792 UP000000803 UP000078542 UP000001292 UP000009192 UP000268350 UP000008744 UP000075883 UP000008711 UP000001819 UP000092445 UP000095300 UP000092460 UP000091820 UP000002282 UP000075900 UP000076408 UP000075903 UP000076407 UP000007062 UP000027135 UP000075901 UP000075881 UP000075880 UP000075885 UP000000673 UP000069272 UP000075886 UP000075882 UP000075840 UP000030765 UP000075884 UP000075902 UP000069940 UP000249989 UP000008820 UP000002320 UP000015103 UP000009046 UP000192223
UP000036403 UP000000311 UP000053097 UP000279307 UP000075809 UP000007755 UP000078540 UP000005205 UP000078492 UP000007798 UP000095301 UP000192221 UP000078541 UP000001070 UP000215335 UP000002358 UP000000304 UP000008792 UP000000803 UP000078542 UP000001292 UP000009192 UP000268350 UP000008744 UP000075883 UP000008711 UP000001819 UP000092445 UP000095300 UP000092460 UP000091820 UP000002282 UP000075900 UP000076408 UP000075903 UP000076407 UP000007062 UP000027135 UP000075901 UP000075881 UP000075880 UP000075885 UP000000673 UP000069272 UP000075886 UP000075882 UP000075840 UP000030765 UP000075884 UP000075902 UP000069940 UP000249989 UP000008820 UP000002320 UP000015103 UP000009046 UP000192223
Pfam
PF07978 NIPSNAP
SUPFAM
SSF54909
SSF54909
ProteinModelPortal
Q2F671
A0A2H1VIT7
A0A2A4JK64
A0A194Q1Q4
A0A194RSL7
A0A212FMJ6
+ More
A0A437BIN3 S4PD61 A0A0L7L306 A0A0J7NWY4 E2ANA8 E9IVX2 A0A026WEC4 A0A151WY94 F4WQ54 A0A151I5Y0 A0A158NX73 A0A195EDJ3 B4NET2 A0A1I8MNN4 A0A1W4UN95 A0A151JU04 B4JLK2 A0A232FBS8 K7J4E3 A0A1W4V295 B4R5Q9 U5EIJ3 A0A1L8EE23 A0A0C9RD66 B4MGB9 M9PJP0 Q9VXK0 A0A151I798 B4IL16 A0A1L8EAD3 A0A0Q9XSZ8 B4L7M8 A0A0Q9XG84 A0A3B0K6P1 B4H134 A0A182M5F8 B3NXF3 Q29GK0 A0A1B6CJG4 D3TMS9 A0A1A9ZLF5 A0A1I8Q4E5 W8BXN9 A0A1B0C564 A0A0A1WUC4 A0A1A9W645 B4PXB5 A0A0K8VWH4 A0A182RC46 A0A034WIA3 A0A0C9R3Y7 A0A2M3Z567 A0A182YHI0 A0A182V8Z3 A0A182XE39 Q7PGV6 A0A067QIR2 A0A2M3Z573 A0A182SCL7 A0A2M4AGT3 A0A0K8TP78 A0A182JWB1 A0A1B6F0A0 A0A2M4BWM2 A0A2M4AGY6 A0A2M4BWF4 A0A1L8DW63 A0A182JIL2 T1DNR2 A0A182PT26 W5JDE4 A0A182FKU5 A0A182PZI3 A0A1B6MD62 A0A182LCF0 A0A182HJ84 A0A084VAK1 A0A182N509 A0A182UBT9 A0A023EM56 A0A2M4ALA4 A0A1Q3FHB1 Q17Q08 A0A1S4EV42 A0A1L8DW49 B0X2Y6 A0A336LQI5 A0A023F895 R4FM21 R4WD15 A0A069DRZ0 E0VR64 A0A1Y1KH08 A0A1W4X9P5
A0A437BIN3 S4PD61 A0A0L7L306 A0A0J7NWY4 E2ANA8 E9IVX2 A0A026WEC4 A0A151WY94 F4WQ54 A0A151I5Y0 A0A158NX73 A0A195EDJ3 B4NET2 A0A1I8MNN4 A0A1W4UN95 A0A151JU04 B4JLK2 A0A232FBS8 K7J4E3 A0A1W4V295 B4R5Q9 U5EIJ3 A0A1L8EE23 A0A0C9RD66 B4MGB9 M9PJP0 Q9VXK0 A0A151I798 B4IL16 A0A1L8EAD3 A0A0Q9XSZ8 B4L7M8 A0A0Q9XG84 A0A3B0K6P1 B4H134 A0A182M5F8 B3NXF3 Q29GK0 A0A1B6CJG4 D3TMS9 A0A1A9ZLF5 A0A1I8Q4E5 W8BXN9 A0A1B0C564 A0A0A1WUC4 A0A1A9W645 B4PXB5 A0A0K8VWH4 A0A182RC46 A0A034WIA3 A0A0C9R3Y7 A0A2M3Z567 A0A182YHI0 A0A182V8Z3 A0A182XE39 Q7PGV6 A0A067QIR2 A0A2M3Z573 A0A182SCL7 A0A2M4AGT3 A0A0K8TP78 A0A182JWB1 A0A1B6F0A0 A0A2M4BWM2 A0A2M4AGY6 A0A2M4BWF4 A0A1L8DW63 A0A182JIL2 T1DNR2 A0A182PT26 W5JDE4 A0A182FKU5 A0A182PZI3 A0A1B6MD62 A0A182LCF0 A0A182HJ84 A0A084VAK1 A0A182N509 A0A182UBT9 A0A023EM56 A0A2M4ALA4 A0A1Q3FHB1 Q17Q08 A0A1S4EV42 A0A1L8DW49 B0X2Y6 A0A336LQI5 A0A023F895 R4FM21 R4WD15 A0A069DRZ0 E0VR64 A0A1Y1KH08 A0A1W4X9P5
PDB
5KAK
E-value=1.11188e-07,
Score=132
Ontologies
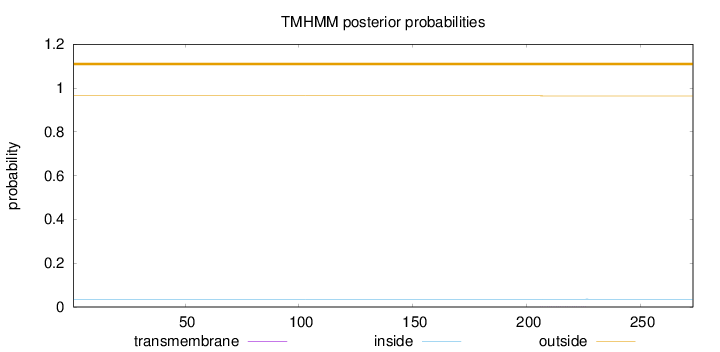
Topology
Length:
273
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00916
Exp number, first 60 AAs:
0.00122
Total prob of N-in:
0.03569
outside
1 - 273
Population Genetic Test Statistics
Pi
285.192734
Theta
215.287814
Tajima's D
0.987479
CLR
0.020672
CSRT
0.654717264136793
Interpretation
Uncertain
