Gene
KWMTBOMO09640
Annotation
hypothetical_protein_KGM_15583_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 1.657 Extracellular Reliability : 1.353
Sequence
CDS
ATGGCTTCCAGCGAGGTGACGAAGCTACAACAACATTTGACGTTGCTAAAGGAAGAGTATGGGAAACTGCAGAGTCATTGTGCTGAGGTAGAGCGAAAGTACACTCTGGCAGCGGCATCCGCCGGCGACCTCAGTGAAACTAGCTTTGTGGCTCGACTTCTAATGACCGTAGCTACACTATATGGTAGAGAAACTTACTCTGACATCAAGATCAAACTGCAAAACAAATCCATGCCGGGACATAAATTTGTTCTGAATGCTCGTTCTGATGACTGGAATGAAGAAGCTCTAAAAGACTTTGATGAATTAGACTGGACGTCTCTACCAGATGATGTAGGGTCAGCCCTATTGAAATGGCTGTACACAGATGTTGTTGATTTAAGTAGAGGAGACTCTTTTGCTTTACAACTGATGAAATCTGCAGCAGGTTTCAAATTGTATGGCCTGGTGAACAAGTGTGAACAAGCTCTCATTGCATCAGTTGGTGTAAGGAGCTGTGTTAAGTTCTATTCAGCAGCAGATGAGATTGGCGCCAACGCACTGAAAGAACACTGTTCAGGACTTATTTCTGCACATTGGGATGATCTAACAGGCGATGATTTCGCGCATATGTCATCAGCGCTTCTCTATCGGATGCTGAAGAGCAAAACCCCGCAACCCCTCCACGGAGCCGTCAGATTGATGCGTGAGGACGTCGTCTTTCTGTGTCTGGTCGAGAATCACGCAAACATGGCGTTGTACTTCTCGAACGTTCTCGATTGTACTAGGTCTTATGATATTCGTTTCTGCTATTCGACGATAGATGGCGTTGCTCTTACCGGCGCCGGCGCGGCGGCGTAA
Protein
MASSEVTKLQQHLTLLKEEYGKLQSHCAEVERKYTLAAASAGDLSETSFVARLLMTVATLYGRETYSDIKIKLQNKSMPGHKFVLNARSDDWNEEALKDFDELDWTSLPDDVGSALLKWLYTDVVDLSRGDSFALQLMKSAAGFKLYGLVNKCEQALIASVGVRSCVKFYSAADEIGANALKEHCSGLISAHWDDLTGDDFAHMSSALLYRMLKSKTPQPLHGAVRLMREDVVFLCLVENHANMALYFSNVLDCTRSYDIRFCYSTIDGVALTGAGAAA
Summary
Uniprot
A0A2A4JJ89
A0A2H1VIX3
A0A212FMI5
A0A194Q690
A0A437BIV6
A0A067RP85
+ More
A0A1I8PJR7 A0A336L854 A0A2S2RB17 A0A2S2R610 A0A2H8THQ5 A0A2S2PD14 J9K6W6 A0A182X392 A0A182TH32 A0A1A9W892 A0A182RH40 A0A336M3S2 A0A182NMP7 A0A182JT60 A0A182IV88 A0A182S842 A0A182PCR7 A0A182HZG8 A0A0L0C737 A0A0K8VIL2 W5J796 A0A182G8T9 A0A182TZ84 B0X1T6 A0A2M3ZI35 A0A182FSW2 A0A1B0DLG6 A0A1A9UN61 A0A1I8M233 T1EAF4 B4GM98 A0A3B0KR40 A0A3B0K231 B5DVQ4 A0A3B0K264 A0A0Q9W842 A0A1B0G6Y2 A0A1B0AA90 A0A1Z5LGQ4 A0A182MTE2 A0A023FV96 B4K4H4 A0A034WLU0 A0A1Q3G2R4 A0A1Z5L507 A0A3F2YX71 A0A1A9XHQ1 A0A1B0B9M1 Q16RW7 A0A182UZ83 A0A182LQH8 B7PTH1 A0A1B6L7N3 Q7QKA9 A0A1B6D0U6 A0A182Y4P8 A0A2M4ADH8 A0A084VB96 A0A2M4BAZ6 A0A2M4BAZ0 A0A2M4CTP2 A0A2M4CTL1 A0A088AFW6 A0A2K6VB32 B4NIW9 A0A0M4EWK4 A0A0K8VC97 A0A0K8VEC1 A0A2A3EGG5 A0A1Y1L0B1 A0A2R5LJ33 A0A131XRG3 A0A0P9A2J0 A0A1L8DIW4 A0A1B6G1J7 A0A2P6KHL5 A0A139WGA2 A0A293LHB9 A0A0A1WKY3 A0A3S3Q2B8 A0A1E1XIC9 A0A0C9RBH5 B4MCC1 A0A0Q9W847 A0A023GFZ3 A0A131XHI5 A0A224YHK2 A0A131YMD8 A0A1S3HUN5 L7MHY9 A0A3L5TTU1 T1J2P3
A0A1I8PJR7 A0A336L854 A0A2S2RB17 A0A2S2R610 A0A2H8THQ5 A0A2S2PD14 J9K6W6 A0A182X392 A0A182TH32 A0A1A9W892 A0A182RH40 A0A336M3S2 A0A182NMP7 A0A182JT60 A0A182IV88 A0A182S842 A0A182PCR7 A0A182HZG8 A0A0L0C737 A0A0K8VIL2 W5J796 A0A182G8T9 A0A182TZ84 B0X1T6 A0A2M3ZI35 A0A182FSW2 A0A1B0DLG6 A0A1A9UN61 A0A1I8M233 T1EAF4 B4GM98 A0A3B0KR40 A0A3B0K231 B5DVQ4 A0A3B0K264 A0A0Q9W842 A0A1B0G6Y2 A0A1B0AA90 A0A1Z5LGQ4 A0A182MTE2 A0A023FV96 B4K4H4 A0A034WLU0 A0A1Q3G2R4 A0A1Z5L507 A0A3F2YX71 A0A1A9XHQ1 A0A1B0B9M1 Q16RW7 A0A182UZ83 A0A182LQH8 B7PTH1 A0A1B6L7N3 Q7QKA9 A0A1B6D0U6 A0A182Y4P8 A0A2M4ADH8 A0A084VB96 A0A2M4BAZ6 A0A2M4BAZ0 A0A2M4CTP2 A0A2M4CTL1 A0A088AFW6 A0A2K6VB32 B4NIW9 A0A0M4EWK4 A0A0K8VC97 A0A0K8VEC1 A0A2A3EGG5 A0A1Y1L0B1 A0A2R5LJ33 A0A131XRG3 A0A0P9A2J0 A0A1L8DIW4 A0A1B6G1J7 A0A2P6KHL5 A0A139WGA2 A0A293LHB9 A0A0A1WKY3 A0A3S3Q2B8 A0A1E1XIC9 A0A0C9RBH5 B4MCC1 A0A0Q9W847 A0A023GFZ3 A0A131XHI5 A0A224YHK2 A0A131YMD8 A0A1S3HUN5 L7MHY9 A0A3L5TTU1 T1J2P3
Pubmed
EMBL
NWSH01001194
PCG72165.1
ODYU01002803
SOQ40751.1
AGBW02007651
OWR54958.1
+ More
KQ459582 KPI98925.1 RSAL01000051 RVE50303.1 KK852507 KDR22440.1 UFQS01002249 UFQT01002249 SSX13599.1 SSX33026.1 GGMS01017986 MBY87189.1 GGMS01015957 MBY85160.1 GFXV01001831 MBW13636.1 GGMR01014217 MBY26836.1 ABLF02034591 ABLF02034600 ABLF02034601 UFQS01000367 UFQT01000367 SSX03217.1 SSX23583.1 APCN01001975 JRES01000826 KNC28057.1 GDHF01013600 JAI38714.1 ADMH02002125 ETN58659.1 JXUM01048240 KQ561553 KXJ78184.1 DS232269 EDS38814.1 GGFM01007432 MBW28183.1 AJVK01016427 AJVK01016428 AJVK01016429 GAMD01001073 JAB00518.1 CH479185 EDW37972.1 OUUW01000014 SPP88356.1 SPP88355.1 CM000070 EDY68462.2 SPP88357.1 CH940657 KRF80956.1 CCAG010013878 GFJQ02000373 JAW06597.1 AXCM01000279 GBBL01002662 JAC24658.1 CH933806 EDW13926.1 GAKP01003630 JAC55322.1 GFDL01000952 JAV34093.1 GFJQ02004692 JAW02278.1 AXCN02001211 JXJN01010490 CH477694 EAT37184.1 ABJB010361842 ABJB010439476 ABJB010900400 ABJB011014244 ABJB011097250 DS785766 EEC09893.1 GEBQ01020346 JAT19631.1 AAAB01008799 EAA03764.3 GEDC01018018 JAS19280.1 GGFK01005357 MBW38678.1 ATLV01006109 ATLV01006110 ATLV01006111 KE524339 KFB35240.1 GGFJ01001020 MBW50161.1 GGFJ01001049 MBW50190.1 GGFL01004457 MBW68635.1 GGFL01004485 MBW68663.1 CH964272 EDW84871.2 CP012526 ALC47942.1 GDHF01015838 JAI36476.1 GDHF01015112 JAI37202.1 KZ288254 PBC30855.1 GEZM01070897 JAV66228.1 GGLE01005424 MBY09550.1 GEFM01007008 GEGO01001835 JAP68788.1 JAR93569.1 CH902623 KPU72688.1 GFDF01007688 JAV06396.1 GECZ01013508 JAS56261.1 MWRG01011721 PRD25809.1 KQ971348 KYB26924.1 GFWV01002538 MAA27268.1 GBXI01015001 JAC99290.1 NCKU01001267 RWS12587.1 GFAC01000437 JAT98751.1 GBYB01013854 JAG83621.1 EDW63203.2 KRF80957.1 GBBM01003515 JAC31903.1 GEFH01002062 JAP66519.1 GFPF01006021 MAA17167.1 GEDV01008440 JAP80117.1 GACK01001459 JAA63575.1 KV585100 OPL33058.1 JH431806
KQ459582 KPI98925.1 RSAL01000051 RVE50303.1 KK852507 KDR22440.1 UFQS01002249 UFQT01002249 SSX13599.1 SSX33026.1 GGMS01017986 MBY87189.1 GGMS01015957 MBY85160.1 GFXV01001831 MBW13636.1 GGMR01014217 MBY26836.1 ABLF02034591 ABLF02034600 ABLF02034601 UFQS01000367 UFQT01000367 SSX03217.1 SSX23583.1 APCN01001975 JRES01000826 KNC28057.1 GDHF01013600 JAI38714.1 ADMH02002125 ETN58659.1 JXUM01048240 KQ561553 KXJ78184.1 DS232269 EDS38814.1 GGFM01007432 MBW28183.1 AJVK01016427 AJVK01016428 AJVK01016429 GAMD01001073 JAB00518.1 CH479185 EDW37972.1 OUUW01000014 SPP88356.1 SPP88355.1 CM000070 EDY68462.2 SPP88357.1 CH940657 KRF80956.1 CCAG010013878 GFJQ02000373 JAW06597.1 AXCM01000279 GBBL01002662 JAC24658.1 CH933806 EDW13926.1 GAKP01003630 JAC55322.1 GFDL01000952 JAV34093.1 GFJQ02004692 JAW02278.1 AXCN02001211 JXJN01010490 CH477694 EAT37184.1 ABJB010361842 ABJB010439476 ABJB010900400 ABJB011014244 ABJB011097250 DS785766 EEC09893.1 GEBQ01020346 JAT19631.1 AAAB01008799 EAA03764.3 GEDC01018018 JAS19280.1 GGFK01005357 MBW38678.1 ATLV01006109 ATLV01006110 ATLV01006111 KE524339 KFB35240.1 GGFJ01001020 MBW50161.1 GGFJ01001049 MBW50190.1 GGFL01004457 MBW68635.1 GGFL01004485 MBW68663.1 CH964272 EDW84871.2 CP012526 ALC47942.1 GDHF01015838 JAI36476.1 GDHF01015112 JAI37202.1 KZ288254 PBC30855.1 GEZM01070897 JAV66228.1 GGLE01005424 MBY09550.1 GEFM01007008 GEGO01001835 JAP68788.1 JAR93569.1 CH902623 KPU72688.1 GFDF01007688 JAV06396.1 GECZ01013508 JAS56261.1 MWRG01011721 PRD25809.1 KQ971348 KYB26924.1 GFWV01002538 MAA27268.1 GBXI01015001 JAC99290.1 NCKU01001267 RWS12587.1 GFAC01000437 JAT98751.1 GBYB01013854 JAG83621.1 EDW63203.2 KRF80957.1 GBBM01003515 JAC31903.1 GEFH01002062 JAP66519.1 GFPF01006021 MAA17167.1 GEDV01008440 JAP80117.1 GACK01001459 JAA63575.1 KV585100 OPL33058.1 JH431806
Proteomes
UP000218220
UP000007151
UP000053268
UP000283053
UP000027135
UP000095300
+ More
UP000007819 UP000076407 UP000075902 UP000091820 UP000075900 UP000075884 UP000075881 UP000075880 UP000075901 UP000075885 UP000075840 UP000037069 UP000000673 UP000069940 UP000249989 UP000002320 UP000069272 UP000092462 UP000078200 UP000095301 UP000008744 UP000268350 UP000001819 UP000008792 UP000092444 UP000092445 UP000075883 UP000009192 UP000075886 UP000092443 UP000092460 UP000008820 UP000075903 UP000075882 UP000001555 UP000007062 UP000076408 UP000030765 UP000005203 UP000007798 UP000092553 UP000242457 UP000007801 UP000007266 UP000285301 UP000085678
UP000007819 UP000076407 UP000075902 UP000091820 UP000075900 UP000075884 UP000075881 UP000075880 UP000075901 UP000075885 UP000075840 UP000037069 UP000000673 UP000069940 UP000249989 UP000002320 UP000069272 UP000092462 UP000078200 UP000095301 UP000008744 UP000268350 UP000001819 UP000008792 UP000092444 UP000092445 UP000075883 UP000009192 UP000075886 UP000092443 UP000092460 UP000008820 UP000075903 UP000075882 UP000001555 UP000007062 UP000076408 UP000030765 UP000005203 UP000007798 UP000092553 UP000242457 UP000007801 UP000007266 UP000285301 UP000085678
PRIDE
Pfam
Interpro
IPR000210
BTB/POZ_dom
+ More
IPR011333 SKP1/BTB/POZ_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR002110 Ankyrin_rpt
IPR011011 Znf_FYVE_PHD
IPR017455 Znf_FYVE-rel
IPR013083 Znf_RING/FYVE/PHD
IPR000306 Znf_FYVE
IPR010625 CHCH
IPR000719 Prot_kinase_dom
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR011333 SKP1/BTB/POZ_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR002110 Ankyrin_rpt
IPR011011 Znf_FYVE_PHD
IPR017455 Znf_FYVE-rel
IPR013083 Znf_RING/FYVE/PHD
IPR000306 Znf_FYVE
IPR010625 CHCH
IPR000719 Prot_kinase_dom
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
Gene 3D
CDD
ProteinModelPortal
A0A2A4JJ89
A0A2H1VIX3
A0A212FMI5
A0A194Q690
A0A437BIV6
A0A067RP85
+ More
A0A1I8PJR7 A0A336L854 A0A2S2RB17 A0A2S2R610 A0A2H8THQ5 A0A2S2PD14 J9K6W6 A0A182X392 A0A182TH32 A0A1A9W892 A0A182RH40 A0A336M3S2 A0A182NMP7 A0A182JT60 A0A182IV88 A0A182S842 A0A182PCR7 A0A182HZG8 A0A0L0C737 A0A0K8VIL2 W5J796 A0A182G8T9 A0A182TZ84 B0X1T6 A0A2M3ZI35 A0A182FSW2 A0A1B0DLG6 A0A1A9UN61 A0A1I8M233 T1EAF4 B4GM98 A0A3B0KR40 A0A3B0K231 B5DVQ4 A0A3B0K264 A0A0Q9W842 A0A1B0G6Y2 A0A1B0AA90 A0A1Z5LGQ4 A0A182MTE2 A0A023FV96 B4K4H4 A0A034WLU0 A0A1Q3G2R4 A0A1Z5L507 A0A3F2YX71 A0A1A9XHQ1 A0A1B0B9M1 Q16RW7 A0A182UZ83 A0A182LQH8 B7PTH1 A0A1B6L7N3 Q7QKA9 A0A1B6D0U6 A0A182Y4P8 A0A2M4ADH8 A0A084VB96 A0A2M4BAZ6 A0A2M4BAZ0 A0A2M4CTP2 A0A2M4CTL1 A0A088AFW6 A0A2K6VB32 B4NIW9 A0A0M4EWK4 A0A0K8VC97 A0A0K8VEC1 A0A2A3EGG5 A0A1Y1L0B1 A0A2R5LJ33 A0A131XRG3 A0A0P9A2J0 A0A1L8DIW4 A0A1B6G1J7 A0A2P6KHL5 A0A139WGA2 A0A293LHB9 A0A0A1WKY3 A0A3S3Q2B8 A0A1E1XIC9 A0A0C9RBH5 B4MCC1 A0A0Q9W847 A0A023GFZ3 A0A131XHI5 A0A224YHK2 A0A131YMD8 A0A1S3HUN5 L7MHY9 A0A3L5TTU1 T1J2P3
A0A1I8PJR7 A0A336L854 A0A2S2RB17 A0A2S2R610 A0A2H8THQ5 A0A2S2PD14 J9K6W6 A0A182X392 A0A182TH32 A0A1A9W892 A0A182RH40 A0A336M3S2 A0A182NMP7 A0A182JT60 A0A182IV88 A0A182S842 A0A182PCR7 A0A182HZG8 A0A0L0C737 A0A0K8VIL2 W5J796 A0A182G8T9 A0A182TZ84 B0X1T6 A0A2M3ZI35 A0A182FSW2 A0A1B0DLG6 A0A1A9UN61 A0A1I8M233 T1EAF4 B4GM98 A0A3B0KR40 A0A3B0K231 B5DVQ4 A0A3B0K264 A0A0Q9W842 A0A1B0G6Y2 A0A1B0AA90 A0A1Z5LGQ4 A0A182MTE2 A0A023FV96 B4K4H4 A0A034WLU0 A0A1Q3G2R4 A0A1Z5L507 A0A3F2YX71 A0A1A9XHQ1 A0A1B0B9M1 Q16RW7 A0A182UZ83 A0A182LQH8 B7PTH1 A0A1B6L7N3 Q7QKA9 A0A1B6D0U6 A0A182Y4P8 A0A2M4ADH8 A0A084VB96 A0A2M4BAZ6 A0A2M4BAZ0 A0A2M4CTP2 A0A2M4CTL1 A0A088AFW6 A0A2K6VB32 B4NIW9 A0A0M4EWK4 A0A0K8VC97 A0A0K8VEC1 A0A2A3EGG5 A0A1Y1L0B1 A0A2R5LJ33 A0A131XRG3 A0A0P9A2J0 A0A1L8DIW4 A0A1B6G1J7 A0A2P6KHL5 A0A139WGA2 A0A293LHB9 A0A0A1WKY3 A0A3S3Q2B8 A0A1E1XIC9 A0A0C9RBH5 B4MCC1 A0A0Q9W847 A0A023GFZ3 A0A131XHI5 A0A224YHK2 A0A131YMD8 A0A1S3HUN5 L7MHY9 A0A3L5TTU1 T1J2P3
Ontologies
GO
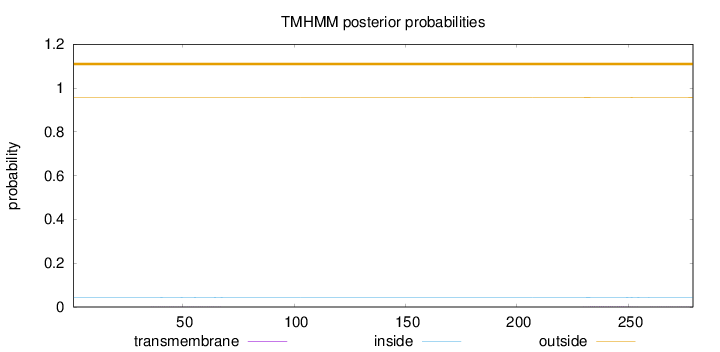
Topology
Length:
279
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0588899999999999
Exp number, first 60 AAs:
0.01491
Total prob of N-in:
0.04300
outside
1 - 279
Population Genetic Test Statistics
Pi
248.157946
Theta
184.173913
Tajima's D
2.266162
CLR
21.40829
CSRT
0.922053897305135
Interpretation
Uncertain
