Gene
KWMTBOMO09639
Annotation
PREDICTED:_uncharacterized_protein_LOC106107543_[Papilio_polytes]
Location in the cell
Extracellular Reliability : 2.801
Sequence
CDS
ATGGAATTAGTCGTATTACTATTAACGTTTGCATCATTTTGGAAAGCCTCGACGAGCTTGCGTTGTTATCAATGCGATCCCGAAGACCCATACAAGACCAGTAACCTGTGCGAAAATTTTGACTACAGCGACAAATTCCTCGTCGAATGTCATTCCTCAACTATGTGCTTCAAGAAACAGACTTACCTACGGGTCGATAATGGAATGAACTCCACGGGAATTCAACGTGGTTGCGCGTCGCAAACTCTGAACGGCGAGCAGAGGAAAATCAATGGAAAATGGCAATACGTTTCGACGATTTACGACGCATACAACGCGACTTGCTTCGAAGACCCGAGTGACTCAGAGAGGGTGACAAAAACTATTTACTGTTACTGCGAGGGAGACAAATGTAACGGAGCTATCCGATTGACCCTCAACTCTTTCTTGGTTCTTATTTTAATTATCATTTCACTAAATTTTACAAGTTAA
Protein
MELVVLLLTFASFWKASTSLRCYQCDPEDPYKTSNLCENFDYSDKFLVECHSSTMCFKKQTYLRVDNGMNSTGIQRGCASQTLNGEQRKINGKWQYVSTIYDAYNATCFEDPSDSERVTKTIYCYCEGDKCNGAIRLTLNSFLVLILIIISLNFTS
Summary
Uniprot
A0A2A4JKN8
A0A194RS88
A0A194Q037
I4DKJ2
A0A2H1VJY6
I4DLG6
+ More
A0A194Q019 A0A194RR36 A0A2A4JL43 A0A1L8DP43 A0A1E1WUT8 A0A139WGE1 A0A2S2NW47 A0A0T6BG90 A0A1B0CGM9 A0A182JCJ8 A0A1Y1LLS9 J9KA85 A0A336M020 A0A0L0C712 A0A0Q9WJ36 E9HRE6 A0A1S4GEA5 A0A182HZG7 A0A2P8Z8U7 A0A034WTM3 A0A0P5E835 A0A0P6CW13 A0A0P5K213 A0A0P4X187 A0A162SBX4 A0A0P5AV11 A0A336MV79 A0A1D2ND17 A0A336N3K7 A0A336MXK6 A0A0P6IEM6 A0A0M4EJV3 A0A182NMP6 A0A0Q9WXM3 A0A226E3K1 A0A182M7P3 A0A0Q9X484 A0A182FSW3 A0A182VQ32 A0A182RH41 B0X1T5 A7UR32 Q16RW6 A0A182X393 A0A182PCR6 A0A182LQH6 A0A182UGW2 A0A182VBF1 A0A1B6JJY7 A0A0K2UW80 A0A182Q9P4 A0A182JT59 A0A1B6MCX8 B4J723 A0A2M4CH47 A0A2M4CH48 A0A0L0C6K8 A0A182G8T8 A0A2S2R8G4 A0A1B6GAA9 A0A1A9UN50 A0A2J7QH37 A0A1I8NJ35 A0A026W3A6 B4LLT5 A0A1A9W880 A0A1J1J0D0 W5J5I1 V9IFG8 Q290D4 A0A088AFT5 N6T0J2 A0A2A3EHB6 B4GCE7 A0A3B0J0K5 B3MCI8 A0A158NBI7 A0A195FCV7 A0A195DRD3
A0A194Q019 A0A194RR36 A0A2A4JL43 A0A1L8DP43 A0A1E1WUT8 A0A139WGE1 A0A2S2NW47 A0A0T6BG90 A0A1B0CGM9 A0A182JCJ8 A0A1Y1LLS9 J9KA85 A0A336M020 A0A0L0C712 A0A0Q9WJ36 E9HRE6 A0A1S4GEA5 A0A182HZG7 A0A2P8Z8U7 A0A034WTM3 A0A0P5E835 A0A0P6CW13 A0A0P5K213 A0A0P4X187 A0A162SBX4 A0A0P5AV11 A0A336MV79 A0A1D2ND17 A0A336N3K7 A0A336MXK6 A0A0P6IEM6 A0A0M4EJV3 A0A182NMP6 A0A0Q9WXM3 A0A226E3K1 A0A182M7P3 A0A0Q9X484 A0A182FSW3 A0A182VQ32 A0A182RH41 B0X1T5 A7UR32 Q16RW6 A0A182X393 A0A182PCR6 A0A182LQH6 A0A182UGW2 A0A182VBF1 A0A1B6JJY7 A0A0K2UW80 A0A182Q9P4 A0A182JT59 A0A1B6MCX8 B4J723 A0A2M4CH47 A0A2M4CH48 A0A0L0C6K8 A0A182G8T8 A0A2S2R8G4 A0A1B6GAA9 A0A1A9UN50 A0A2J7QH37 A0A1I8NJ35 A0A026W3A6 B4LLT5 A0A1A9W880 A0A1J1J0D0 W5J5I1 V9IFG8 Q290D4 A0A088AFT5 N6T0J2 A0A2A3EHB6 B4GCE7 A0A3B0J0K5 B3MCI8 A0A158NBI7 A0A195FCV7 A0A195DRD3
Pubmed
EMBL
NWSH01001194
PCG72164.1
KQ459700
KPJ20342.1
KQ459582
KPI98926.1
+ More
AK401810 BAM18432.1 ODYU01002803 SOQ40752.1 AK402134 BAM18756.1 KPI98927.1 KPJ20343.1 PCG72163.1 GFDF01005875 JAV08209.1 GDQN01000493 JAT90561.1 KQ971348 KYB26925.1 GGMR01008820 MBY21439.1 LJIG01000753 KRT86187.1 AJWK01011468 GEZM01053969 JAV73891.1 ABLF02034591 ABLF02034600 ABLF02034601 UFQT01000367 SSX23584.1 JRES01000826 KNC28060.1 CH940657 KRF80958.1 GL732734 EFX65702.1 AAAB01008799 APCN01001975 PYGN01000144 PSN52921.1 GAKP01000958 JAC57994.1 GDIP01145260 JAJ78142.1 GDIQ01086919 JAN07818.1 GDIQ01192077 JAK59648.1 GDIP01250688 JAI72713.1 LRGB01000056 KZS21165.1 GDIP01197605 JAJ25797.1 UFQT01002249 SSX33029.1 LJIJ01000091 ODN02975.1 SSX33028.1 SSX33027.1 GDIQ01005657 JAN89080.1 CP012524 ALC41592.1 CH933806 KRG00731.1 LNIX01000007 OXA52312.1 AXCM01000279 KRG00730.1 DS232269 EDS38813.1 EDO64816.2 CH477694 EAT37182.1 EAT37183.1 GECU01008211 GECU01000329 JAS99495.1 JAT07378.1 HACA01024974 CDW42335.1 AXCN02001211 GEBQ01006202 JAT33775.1 CH916367 EDW01011.1 GGFL01000489 MBW64667.1 GGFL01000488 MBW64666.1 KNC28053.1 JXUM01048237 JXUM01048238 JXUM01048239 KQ561553 KXJ78183.1 GGMS01017010 MBY86213.1 GECZ01010407 JAS59362.1 NEVH01014358 PNF27889.1 KK107453 EZA50560.1 CH940648 EDW61958.1 CVRI01000066 CRL05855.1 ADMH02002125 ETN58658.1 JR044292 AEY59863.1 CM000071 EAL25429.2 APGK01049600 KB741156 KB632169 ENN73574.1 ERL89489.1 KZ288254 PBC30854.1 CH479181 EDW32424.1 OUUW01000001 SPP74197.1 CH902619 EDV36222.1 ADTU01011113 KQ981673 KYN38243.1 KQ980581 KYN15401.1
AK401810 BAM18432.1 ODYU01002803 SOQ40752.1 AK402134 BAM18756.1 KPI98927.1 KPJ20343.1 PCG72163.1 GFDF01005875 JAV08209.1 GDQN01000493 JAT90561.1 KQ971348 KYB26925.1 GGMR01008820 MBY21439.1 LJIG01000753 KRT86187.1 AJWK01011468 GEZM01053969 JAV73891.1 ABLF02034591 ABLF02034600 ABLF02034601 UFQT01000367 SSX23584.1 JRES01000826 KNC28060.1 CH940657 KRF80958.1 GL732734 EFX65702.1 AAAB01008799 APCN01001975 PYGN01000144 PSN52921.1 GAKP01000958 JAC57994.1 GDIP01145260 JAJ78142.1 GDIQ01086919 JAN07818.1 GDIQ01192077 JAK59648.1 GDIP01250688 JAI72713.1 LRGB01000056 KZS21165.1 GDIP01197605 JAJ25797.1 UFQT01002249 SSX33029.1 LJIJ01000091 ODN02975.1 SSX33028.1 SSX33027.1 GDIQ01005657 JAN89080.1 CP012524 ALC41592.1 CH933806 KRG00731.1 LNIX01000007 OXA52312.1 AXCM01000279 KRG00730.1 DS232269 EDS38813.1 EDO64816.2 CH477694 EAT37182.1 EAT37183.1 GECU01008211 GECU01000329 JAS99495.1 JAT07378.1 HACA01024974 CDW42335.1 AXCN02001211 GEBQ01006202 JAT33775.1 CH916367 EDW01011.1 GGFL01000489 MBW64667.1 GGFL01000488 MBW64666.1 KNC28053.1 JXUM01048237 JXUM01048238 JXUM01048239 KQ561553 KXJ78183.1 GGMS01017010 MBY86213.1 GECZ01010407 JAS59362.1 NEVH01014358 PNF27889.1 KK107453 EZA50560.1 CH940648 EDW61958.1 CVRI01000066 CRL05855.1 ADMH02002125 ETN58658.1 JR044292 AEY59863.1 CM000071 EAL25429.2 APGK01049600 KB741156 KB632169 ENN73574.1 ERL89489.1 KZ288254 PBC30854.1 CH479181 EDW32424.1 OUUW01000001 SPP74197.1 CH902619 EDV36222.1 ADTU01011113 KQ981673 KYN38243.1 KQ980581 KYN15401.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007266
UP000092461
UP000075880
+ More
UP000007819 UP000037069 UP000008792 UP000000305 UP000075840 UP000245037 UP000076858 UP000094527 UP000092553 UP000075884 UP000009192 UP000198287 UP000075883 UP000069272 UP000075920 UP000075900 UP000002320 UP000007062 UP000008820 UP000076407 UP000075885 UP000075882 UP000075902 UP000075903 UP000075886 UP000075881 UP000001070 UP000069940 UP000249989 UP000078200 UP000235965 UP000095301 UP000053097 UP000091820 UP000183832 UP000000673 UP000001819 UP000005203 UP000019118 UP000030742 UP000242457 UP000008744 UP000268350 UP000007801 UP000005205 UP000078541 UP000078492
UP000007819 UP000037069 UP000008792 UP000000305 UP000075840 UP000245037 UP000076858 UP000094527 UP000092553 UP000075884 UP000009192 UP000198287 UP000075883 UP000069272 UP000075920 UP000075900 UP000002320 UP000007062 UP000008820 UP000076407 UP000075885 UP000075882 UP000075902 UP000075903 UP000075886 UP000075881 UP000001070 UP000069940 UP000249989 UP000078200 UP000235965 UP000095301 UP000053097 UP000091820 UP000183832 UP000000673 UP000001819 UP000005203 UP000019118 UP000030742 UP000242457 UP000008744 UP000268350 UP000007801 UP000005205 UP000078541 UP000078492
Pfam
PF00021 UPAR_LY6
Gene 3D
ProteinModelPortal
A0A2A4JKN8
A0A194RS88
A0A194Q037
I4DKJ2
A0A2H1VJY6
I4DLG6
+ More
A0A194Q019 A0A194RR36 A0A2A4JL43 A0A1L8DP43 A0A1E1WUT8 A0A139WGE1 A0A2S2NW47 A0A0T6BG90 A0A1B0CGM9 A0A182JCJ8 A0A1Y1LLS9 J9KA85 A0A336M020 A0A0L0C712 A0A0Q9WJ36 E9HRE6 A0A1S4GEA5 A0A182HZG7 A0A2P8Z8U7 A0A034WTM3 A0A0P5E835 A0A0P6CW13 A0A0P5K213 A0A0P4X187 A0A162SBX4 A0A0P5AV11 A0A336MV79 A0A1D2ND17 A0A336N3K7 A0A336MXK6 A0A0P6IEM6 A0A0M4EJV3 A0A182NMP6 A0A0Q9WXM3 A0A226E3K1 A0A182M7P3 A0A0Q9X484 A0A182FSW3 A0A182VQ32 A0A182RH41 B0X1T5 A7UR32 Q16RW6 A0A182X393 A0A182PCR6 A0A182LQH6 A0A182UGW2 A0A182VBF1 A0A1B6JJY7 A0A0K2UW80 A0A182Q9P4 A0A182JT59 A0A1B6MCX8 B4J723 A0A2M4CH47 A0A2M4CH48 A0A0L0C6K8 A0A182G8T8 A0A2S2R8G4 A0A1B6GAA9 A0A1A9UN50 A0A2J7QH37 A0A1I8NJ35 A0A026W3A6 B4LLT5 A0A1A9W880 A0A1J1J0D0 W5J5I1 V9IFG8 Q290D4 A0A088AFT5 N6T0J2 A0A2A3EHB6 B4GCE7 A0A3B0J0K5 B3MCI8 A0A158NBI7 A0A195FCV7 A0A195DRD3
A0A194Q019 A0A194RR36 A0A2A4JL43 A0A1L8DP43 A0A1E1WUT8 A0A139WGE1 A0A2S2NW47 A0A0T6BG90 A0A1B0CGM9 A0A182JCJ8 A0A1Y1LLS9 J9KA85 A0A336M020 A0A0L0C712 A0A0Q9WJ36 E9HRE6 A0A1S4GEA5 A0A182HZG7 A0A2P8Z8U7 A0A034WTM3 A0A0P5E835 A0A0P6CW13 A0A0P5K213 A0A0P4X187 A0A162SBX4 A0A0P5AV11 A0A336MV79 A0A1D2ND17 A0A336N3K7 A0A336MXK6 A0A0P6IEM6 A0A0M4EJV3 A0A182NMP6 A0A0Q9WXM3 A0A226E3K1 A0A182M7P3 A0A0Q9X484 A0A182FSW3 A0A182VQ32 A0A182RH41 B0X1T5 A7UR32 Q16RW6 A0A182X393 A0A182PCR6 A0A182LQH6 A0A182UGW2 A0A182VBF1 A0A1B6JJY7 A0A0K2UW80 A0A182Q9P4 A0A182JT59 A0A1B6MCX8 B4J723 A0A2M4CH47 A0A2M4CH48 A0A0L0C6K8 A0A182G8T8 A0A2S2R8G4 A0A1B6GAA9 A0A1A9UN50 A0A2J7QH37 A0A1I8NJ35 A0A026W3A6 B4LLT5 A0A1A9W880 A0A1J1J0D0 W5J5I1 V9IFG8 Q290D4 A0A088AFT5 N6T0J2 A0A2A3EHB6 B4GCE7 A0A3B0J0K5 B3MCI8 A0A158NBI7 A0A195FCV7 A0A195DRD3
Ontologies
GO
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.998318
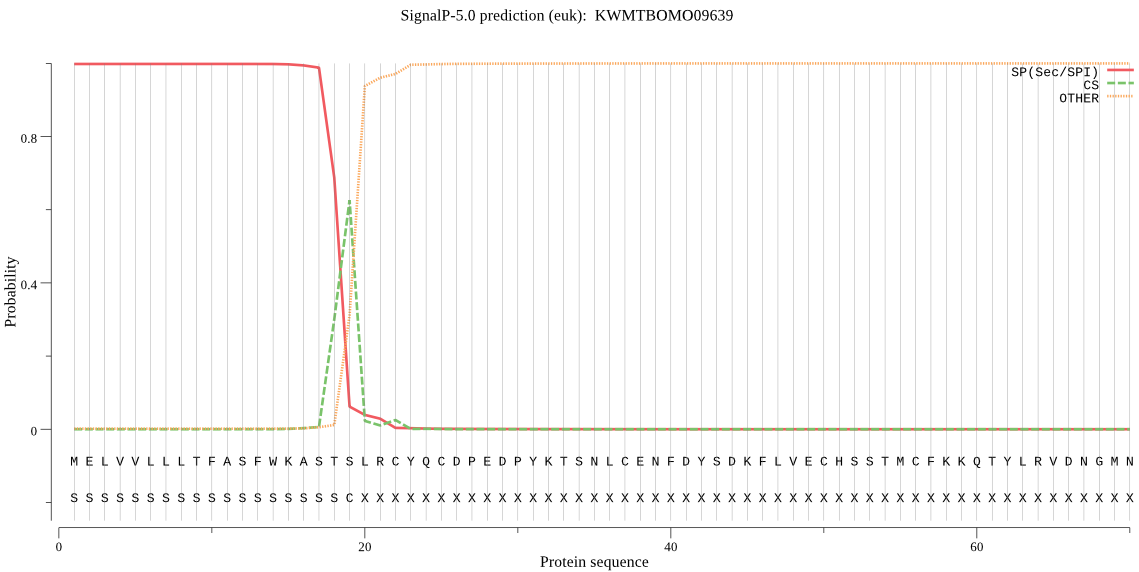
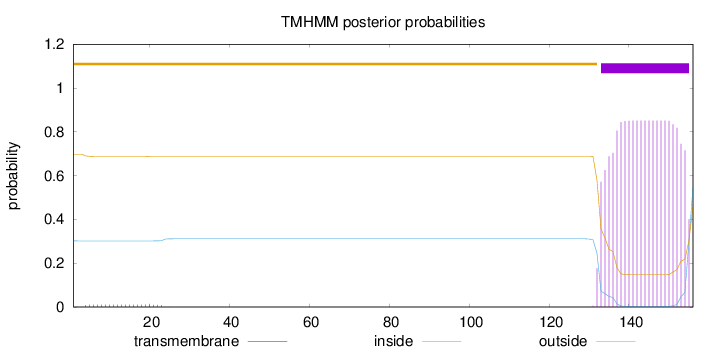
Length:
156
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.35409
Exp number, first 60 AAs:
0.20953
Total prob of N-in:
0.30183
outside
1 - 132
TMhelix
133 - 155
inside
156 - 156
Population Genetic Test Statistics
Pi
232.214729
Theta
169.296385
Tajima's D
1.352774
CLR
0.020191
CSRT
0.752312384380781
Interpretation
Uncertain
