Gene
KWMTBOMO09636 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012857
Annotation
PREDICTED:_cytochrome_b-c1_complex_subunit_6?_mitochondrial-like_[Papilio_xuthus]
Full name
Cytochrome b-c1 complex subunit 6
Location in the cell
Nuclear Reliability : 2.719
Sequence
CDS
ATGTCTGACAAATTCGTTCCCGTGGTTAAGGCAGAGGAAACAGAGGAGGAGGAGTTGGTGGATCCGCAGCAGTCTCTTAGGGAAGAGTGTTCCCAAAAGCCTGATGCCCAAAACATGTGGGCTAAATATCAAGAGTGCAATGATCGTGTAAACTCCAGATCTAAAACTGCAGAAACCTGTGAAGAAGAACTCATTGACTATGTCCATGTTCTTGACAAATGCGTCACCAAGGACTTGTTTAAGAGGCTAAAGTAA
Protein
MSDKFVPVVKAEETEEEELVDPQQSLREECSQKPDAQNMWAKYQECNDRVNSRSKTAETCEEELIDYVHVLDKCVTKDLFKRLK
Summary
Description
This is a component of the ubiquinol-cytochrome c reductase complex (complex III or cytochrome b-c1 complex), which is part of the mitochondrial respiratory chain. This protein may mediate formation of the complex between cytochromes c and c1.
Similarity
Belongs to the UQCRH/QCR6 family.
Feature
chain Cytochrome b-c1 complex subunit 6
Uniprot
H9JTJ5
I4DLG5
A0A194RSM2
A0A3S2NW40
A0A212FMK8
A0A2H1V8E7
+ More
A0A2A4IUL1 Q8I918 A0A1J1J0J6 A0A1L8E172 A0A1L8E1M2 A0A182JT57 A0A182TRP9 A0A182PCR4 A0A084VB95 A0A182UTD1 A0A182X395 Q7QKB0 A0A182HZG5 A0A182RH43 A0A182J7M4 A0A1B0CI53 A0A182G8T6 A0A023EDZ6 A0A182M5A1 A0A182Y4P9 D2A452 A0A0M5J1V2 A0A1L8EIF1 A0A026W4P0 Q1HR18 J9HJ41 A0A2J7QH09 A0A0K8TTF0 A0A182VQ30 T1DDV3 A0A3B0K295 B4NIW8 A0A182NMP4 A0A182FSW5 B4LVY4 F4X8N7 A0A0P8XD53 A0A0Q5WBY1 A0A182QFR2 B3MTL1 A0A0L0C744 B5DVQ3 A0A2M3Z8R6 T1E1V6 W8CBP8 B4GM97 A0A1I8MK27 T1DPZ2 A0A2M4AE75 A0A2M4C4R1 A0A2M4C5V4 A0A1W4UKB7 A0A1Q3FKW2 B3P244 W5J465 B4K4H5 B0WLN6 B4PT18 A0A1A9W882 A0A0R1DPD7 A0A1A9UN43 A0A336LZS3 A0A1B0AA60 A0A0M5J7P8 A0A1I8P257 A0A1A9XHP9 A0A1B0B9M0 A0A0J9R856 Q4QPY6 B4ILS1 B4NTK4 B4JYT5 A0A2P8Z8V5 A0A151IGX8 J3JVD7 L7MKY7 A0A195BAK9 A0A158NBI8 A8Y535 A0A1W4W0S7 D2NUK7 E2A9F7 N6SVI7 A0A0L7RER5 A0A195DSA3 A0A195FDZ1 A0A131XJY4 A0A067RRN8 A0A0J7KQ30 A0A1Y1N2F7 A0A2A3EHV8 E2C3Y6 A0A088AFT4 A0A1V9XQ00
A0A2A4IUL1 Q8I918 A0A1J1J0J6 A0A1L8E172 A0A1L8E1M2 A0A182JT57 A0A182TRP9 A0A182PCR4 A0A084VB95 A0A182UTD1 A0A182X395 Q7QKB0 A0A182HZG5 A0A182RH43 A0A182J7M4 A0A1B0CI53 A0A182G8T6 A0A023EDZ6 A0A182M5A1 A0A182Y4P9 D2A452 A0A0M5J1V2 A0A1L8EIF1 A0A026W4P0 Q1HR18 J9HJ41 A0A2J7QH09 A0A0K8TTF0 A0A182VQ30 T1DDV3 A0A3B0K295 B4NIW8 A0A182NMP4 A0A182FSW5 B4LVY4 F4X8N7 A0A0P8XD53 A0A0Q5WBY1 A0A182QFR2 B3MTL1 A0A0L0C744 B5DVQ3 A0A2M3Z8R6 T1E1V6 W8CBP8 B4GM97 A0A1I8MK27 T1DPZ2 A0A2M4AE75 A0A2M4C4R1 A0A2M4C5V4 A0A1W4UKB7 A0A1Q3FKW2 B3P244 W5J465 B4K4H5 B0WLN6 B4PT18 A0A1A9W882 A0A0R1DPD7 A0A1A9UN43 A0A336LZS3 A0A1B0AA60 A0A0M5J7P8 A0A1I8P257 A0A1A9XHP9 A0A1B0B9M0 A0A0J9R856 Q4QPY6 B4ILS1 B4NTK4 B4JYT5 A0A2P8Z8V5 A0A151IGX8 J3JVD7 L7MKY7 A0A195BAK9 A0A158NBI8 A8Y535 A0A1W4W0S7 D2NUK7 E2A9F7 N6SVI7 A0A0L7RER5 A0A195DSA3 A0A195FDZ1 A0A131XJY4 A0A067RRN8 A0A0J7KQ30 A0A1Y1N2F7 A0A2A3EHV8 E2C3Y6 A0A088AFT4 A0A1V9XQ00
Pubmed
19121390
22651552
26354079
22118469
24438588
12364791
+ More
14747013 17210077 26483478 24945155 25244985 18362917 19820115 24508170 30249741 17204158 17510324 26369729 24330624 17994087 21719571 26108605 15632085 23185243 24495485 25315136 20920257 23761445 18057021 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 22516182 25576852 21347285 20798317 23537049 28049606 24845553 28004739 28327890
14747013 17210077 26483478 24945155 25244985 18362917 19820115 24508170 30249741 17204158 17510324 26369729 24330624 17994087 21719571 26108605 15632085 23185243 24495485 25315136 20920257 23761445 18057021 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 22516182 25576852 21347285 20798317 23537049 28049606 24845553 28004739 28327890
EMBL
BABH01003695
AK402133
KQ459582
BAM18755.1
KPI98930.1
KQ459700
+ More
KPJ20345.1 RSAL01000051 RVE50302.1 AGBW02007651 OWR54963.1 ODYU01001199 SOQ37087.1 NWSH01006093 PCG63667.1 AY071867 EF635220 AAL60593.1 ABU96711.1 CVRI01000066 CRL05856.1 GFDF01001688 JAV12396.1 GFDF01001679 JAV12405.1 ATLV01006109 KE524339 KFB35239.1 AAAB01008799 EAA03739.5 APCN01001975 AJWK01013041 JXUM01043531 JXUM01048232 KQ561553 KQ561365 KXJ78181.1 KXJ78836.1 GAPW01006582 JAC07016.1 AXCM01000279 KQ971348 EFA05661.1 CP012523 ALC39711.1 GFDG01000365 JAV18434.1 KK107453 QOIP01000004 EZA50561.1 RLU23452.1 DQ440276 CH477694 ABF18309.1 EAT37180.1 EJY57884.1 NEVH01014358 PNF27867.1 GDAI01000167 JAI17436.1 GALA01001307 JAA93545.1 OUUW01000014 SPP88354.1 CH964272 EDW84870.1 CH940650 EDW66489.1 GL888932 EGI57306.1 CH902623 KPU72689.1 CH954177 KQS70870.1 AXCN02001211 EDV30142.1 JRES01000826 KNC28061.1 CM000070 EDY68461.1 GGFM01004141 MBW24892.1 GALA01001334 JAA93518.1 GAMC01000304 JAC06252.1 CH479185 EDW37971.1 GAMD01002904 JAA98686.1 GGFK01005729 MBW39050.1 GGFJ01011205 MBW60346.1 GGFJ01011207 MBW60348.1 GFDL01006795 JAV28250.1 CH954181 EDV47722.1 ADMH02002125 ETN58656.1 CH933806 EDW13927.1 KRG00732.1 DS231988 EDS30578.1 CM000160 EDW95607.1 CM000157 KRJ98381.1 UFQT01000367 SSX23586.1 CP012526 ALC47944.1 JXJN01010481 CM002911 KMY92312.1 AE013599 BT023630 KX531341 AAM68831.1 AAY85030.1 ANY27151.1 CH480886 EDW55363.1 CH983192 CM002910 CM002912 EDX15662.1 KMY91453.1 KMY91536.1 KMY91539.1 KMY91543.1 KMZ01322.1 CH916378 EDV98550.1 PYGN01000144 PSN52930.1 KQ977661 KYN00703.1 BT127204 AEE62166.1 GACK01001086 JAA63948.1 KQ976540 KYM81255.1 ADTU01011115 AE014296 EDP28072.2 BT120150 ADB57071.1 GL437855 EFN69930.1 APGK01054966 APGK01054970 KB741256 ENN71745.1 ENN71759.1 KQ414609 KOC69333.1 KQ980581 KYN15399.1 KQ981673 KYN38244.1 GEFH01002076 JAP66505.1 KK852507 KDR22439.1 LBMM01004467 KMQ92386.1 GEZM01014679 JAV92042.1 KZ288254 PBC30852.1 GL452364 EFN77372.1 MNPL01006170 OQR75574.1
KPJ20345.1 RSAL01000051 RVE50302.1 AGBW02007651 OWR54963.1 ODYU01001199 SOQ37087.1 NWSH01006093 PCG63667.1 AY071867 EF635220 AAL60593.1 ABU96711.1 CVRI01000066 CRL05856.1 GFDF01001688 JAV12396.1 GFDF01001679 JAV12405.1 ATLV01006109 KE524339 KFB35239.1 AAAB01008799 EAA03739.5 APCN01001975 AJWK01013041 JXUM01043531 JXUM01048232 KQ561553 KQ561365 KXJ78181.1 KXJ78836.1 GAPW01006582 JAC07016.1 AXCM01000279 KQ971348 EFA05661.1 CP012523 ALC39711.1 GFDG01000365 JAV18434.1 KK107453 QOIP01000004 EZA50561.1 RLU23452.1 DQ440276 CH477694 ABF18309.1 EAT37180.1 EJY57884.1 NEVH01014358 PNF27867.1 GDAI01000167 JAI17436.1 GALA01001307 JAA93545.1 OUUW01000014 SPP88354.1 CH964272 EDW84870.1 CH940650 EDW66489.1 GL888932 EGI57306.1 CH902623 KPU72689.1 CH954177 KQS70870.1 AXCN02001211 EDV30142.1 JRES01000826 KNC28061.1 CM000070 EDY68461.1 GGFM01004141 MBW24892.1 GALA01001334 JAA93518.1 GAMC01000304 JAC06252.1 CH479185 EDW37971.1 GAMD01002904 JAA98686.1 GGFK01005729 MBW39050.1 GGFJ01011205 MBW60346.1 GGFJ01011207 MBW60348.1 GFDL01006795 JAV28250.1 CH954181 EDV47722.1 ADMH02002125 ETN58656.1 CH933806 EDW13927.1 KRG00732.1 DS231988 EDS30578.1 CM000160 EDW95607.1 CM000157 KRJ98381.1 UFQT01000367 SSX23586.1 CP012526 ALC47944.1 JXJN01010481 CM002911 KMY92312.1 AE013599 BT023630 KX531341 AAM68831.1 AAY85030.1 ANY27151.1 CH480886 EDW55363.1 CH983192 CM002910 CM002912 EDX15662.1 KMY91453.1 KMY91536.1 KMY91539.1 KMY91543.1 KMZ01322.1 CH916378 EDV98550.1 PYGN01000144 PSN52930.1 KQ977661 KYN00703.1 BT127204 AEE62166.1 GACK01001086 JAA63948.1 KQ976540 KYM81255.1 ADTU01011115 AE014296 EDP28072.2 BT120150 ADB57071.1 GL437855 EFN69930.1 APGK01054966 APGK01054970 KB741256 ENN71745.1 ENN71759.1 KQ414609 KOC69333.1 KQ980581 KYN15399.1 KQ981673 KYN38244.1 GEFH01002076 JAP66505.1 KK852507 KDR22439.1 LBMM01004467 KMQ92386.1 GEZM01014679 JAV92042.1 KZ288254 PBC30852.1 GL452364 EFN77372.1 MNPL01006170 OQR75574.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000283053
UP000007151
UP000218220
+ More
UP000183832 UP000075881 UP000075902 UP000075885 UP000030765 UP000075903 UP000076407 UP000007062 UP000075840 UP000075900 UP000075880 UP000092461 UP000069940 UP000249989 UP000075883 UP000076408 UP000007266 UP000092553 UP000053097 UP000279307 UP000008820 UP000235965 UP000075920 UP000268350 UP000007798 UP000075884 UP000069272 UP000008792 UP000007755 UP000007801 UP000008711 UP000075886 UP000037069 UP000001819 UP000008744 UP000095301 UP000192221 UP000000673 UP000009192 UP000002320 UP000002282 UP000091820 UP000078200 UP000092445 UP000095300 UP000092443 UP000092460 UP000000803 UP000001292 UP000000304 UP000001070 UP000245037 UP000078542 UP000078540 UP000005205 UP000000311 UP000019118 UP000053825 UP000078492 UP000078541 UP000027135 UP000036403 UP000242457 UP000008237 UP000005203 UP000192247
UP000183832 UP000075881 UP000075902 UP000075885 UP000030765 UP000075903 UP000076407 UP000007062 UP000075840 UP000075900 UP000075880 UP000092461 UP000069940 UP000249989 UP000075883 UP000076408 UP000007266 UP000092553 UP000053097 UP000279307 UP000008820 UP000235965 UP000075920 UP000268350 UP000007798 UP000075884 UP000069272 UP000008792 UP000007755 UP000007801 UP000008711 UP000075886 UP000037069 UP000001819 UP000008744 UP000095301 UP000192221 UP000000673 UP000009192 UP000002320 UP000002282 UP000091820 UP000078200 UP000092445 UP000095300 UP000092443 UP000092460 UP000000803 UP000001292 UP000000304 UP000001070 UP000245037 UP000078542 UP000078540 UP000005205 UP000000311 UP000019118 UP000053825 UP000078492 UP000078541 UP000027135 UP000036403 UP000242457 UP000008237 UP000005203 UP000192247
Pfam
PF02320 UCR_hinge
Interpro
SUPFAM
SSF81531
SSF81531
Gene 3D
ProteinModelPortal
H9JTJ5
I4DLG5
A0A194RSM2
A0A3S2NW40
A0A212FMK8
A0A2H1V8E7
+ More
A0A2A4IUL1 Q8I918 A0A1J1J0J6 A0A1L8E172 A0A1L8E1M2 A0A182JT57 A0A182TRP9 A0A182PCR4 A0A084VB95 A0A182UTD1 A0A182X395 Q7QKB0 A0A182HZG5 A0A182RH43 A0A182J7M4 A0A1B0CI53 A0A182G8T6 A0A023EDZ6 A0A182M5A1 A0A182Y4P9 D2A452 A0A0M5J1V2 A0A1L8EIF1 A0A026W4P0 Q1HR18 J9HJ41 A0A2J7QH09 A0A0K8TTF0 A0A182VQ30 T1DDV3 A0A3B0K295 B4NIW8 A0A182NMP4 A0A182FSW5 B4LVY4 F4X8N7 A0A0P8XD53 A0A0Q5WBY1 A0A182QFR2 B3MTL1 A0A0L0C744 B5DVQ3 A0A2M3Z8R6 T1E1V6 W8CBP8 B4GM97 A0A1I8MK27 T1DPZ2 A0A2M4AE75 A0A2M4C4R1 A0A2M4C5V4 A0A1W4UKB7 A0A1Q3FKW2 B3P244 W5J465 B4K4H5 B0WLN6 B4PT18 A0A1A9W882 A0A0R1DPD7 A0A1A9UN43 A0A336LZS3 A0A1B0AA60 A0A0M5J7P8 A0A1I8P257 A0A1A9XHP9 A0A1B0B9M0 A0A0J9R856 Q4QPY6 B4ILS1 B4NTK4 B4JYT5 A0A2P8Z8V5 A0A151IGX8 J3JVD7 L7MKY7 A0A195BAK9 A0A158NBI8 A8Y535 A0A1W4W0S7 D2NUK7 E2A9F7 N6SVI7 A0A0L7RER5 A0A195DSA3 A0A195FDZ1 A0A131XJY4 A0A067RRN8 A0A0J7KQ30 A0A1Y1N2F7 A0A2A3EHV8 E2C3Y6 A0A088AFT4 A0A1V9XQ00
A0A2A4IUL1 Q8I918 A0A1J1J0J6 A0A1L8E172 A0A1L8E1M2 A0A182JT57 A0A182TRP9 A0A182PCR4 A0A084VB95 A0A182UTD1 A0A182X395 Q7QKB0 A0A182HZG5 A0A182RH43 A0A182J7M4 A0A1B0CI53 A0A182G8T6 A0A023EDZ6 A0A182M5A1 A0A182Y4P9 D2A452 A0A0M5J1V2 A0A1L8EIF1 A0A026W4P0 Q1HR18 J9HJ41 A0A2J7QH09 A0A0K8TTF0 A0A182VQ30 T1DDV3 A0A3B0K295 B4NIW8 A0A182NMP4 A0A182FSW5 B4LVY4 F4X8N7 A0A0P8XD53 A0A0Q5WBY1 A0A182QFR2 B3MTL1 A0A0L0C744 B5DVQ3 A0A2M3Z8R6 T1E1V6 W8CBP8 B4GM97 A0A1I8MK27 T1DPZ2 A0A2M4AE75 A0A2M4C4R1 A0A2M4C5V4 A0A1W4UKB7 A0A1Q3FKW2 B3P244 W5J465 B4K4H5 B0WLN6 B4PT18 A0A1A9W882 A0A0R1DPD7 A0A1A9UN43 A0A336LZS3 A0A1B0AA60 A0A0M5J7P8 A0A1I8P257 A0A1A9XHP9 A0A1B0B9M0 A0A0J9R856 Q4QPY6 B4ILS1 B4NTK4 B4JYT5 A0A2P8Z8V5 A0A151IGX8 J3JVD7 L7MKY7 A0A195BAK9 A0A158NBI8 A8Y535 A0A1W4W0S7 D2NUK7 E2A9F7 N6SVI7 A0A0L7RER5 A0A195DSA3 A0A195FDZ1 A0A131XJY4 A0A067RRN8 A0A0J7KQ30 A0A1Y1N2F7 A0A2A3EHV8 E2C3Y6 A0A088AFT4 A0A1V9XQ00
PDB
4U3F
E-value=2.16704e-08,
Score=133
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Mitochondrion inner membrane
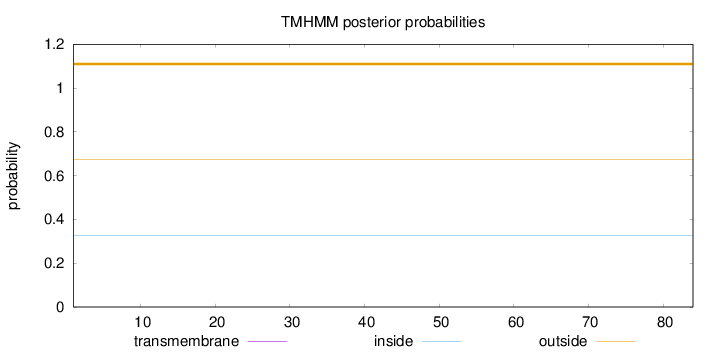
Length:
84
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.32672
outside
1 - 84
Population Genetic Test Statistics
Pi
272.909987
Theta
199.330225
Tajima's D
1.229698
CLR
0.765779
CSRT
0.71966401679916
Interpretation
Uncertain
