Pre Gene Modal
BGIBMGA012856
Annotation
hypothetical_protein_KGM_15471_[Danaus_plexippus]
Full name
Citron Rho-interacting kinase
+ More
Non-specific serine/threonine protein kinase
Non-specific serine/threonine protein kinase
Alternative Name
Rho-interacting, serine/threonine-protein kinase 21
Serine/threonine-protein kinase 21
Serine/threonine-protein kinase 21
Location in the cell
Nuclear Reliability : 2.47
Sequence
CDS
ATGGAACCTTCAAAGGAAGCAATAGCTGTGCGTATAGCACGATTAAATCGACAAATACTAGGCAAAGCAACAAGTGGTGTAAGTAGAAGCAAGAAAACAGTGGACCGAGAAGCATTGTTGGACGCACTTACAGTATTATACGATGAATGTAACGATGATCCTATCAAGAAAACTGATGAATTGGTTCGGGCTTTTGTGGATAAATACCGCAGCACTTTGGCAGAACTGCGAAGGACTAGAGTTTGTATTTCTGACTTTGAAGTAGTTCAAACCATTGGACGTGGATATTTTGGGGAAGTAAATATGGTGAGAGAAAAACAAAGTGGTGATGTATATGCCCTAAAAACATTACGCAAGGAACAATCACGGAAACGTGCCATTGCTTGCACTGAAGATGAGAGGGATGTGTTGGCATCAACGACTAGTCCTTGGATCCCACGTTTGCAGTATGCTTTTCAGGATAGCAACAATTTATACTTAGTAATGGAGCTATGTTCCGGTGGAGATCTTGCCGGGCTGATGTCTCGTCGAAACCATCCATTATCAGAACGAGATGCAGCATTCTACATTGCCGAAGTTGCACATGCTCTGAAAGCACTGCATGGGATGGGTTACATACATAGAGATGTCAAGCCCCACAACATATTTATTGATAGGTAA
Protein
MEPSKEAIAVRIARLNRQILGKATSGVSRSKKTVDREALLDALTVLYDECNDDPIKKTDELVRAFVDKYRSTLAELRRTRVCISDFEVVQTIGRGYFGEVNMVREKQSGDVYALKTLRKEQSRKRAIACTEDERDVLASTTSPWIPRLQYAFQDSNNLYLVMELCSGGDLAGLMSRRNHPLSERDAAFYIAEVAHALKALHGMGYIHRDVKPHNIFIDR
Summary
Description
Plays a role in cytokinesis. Required for KIF14 localization to the central spindle and midbody. Probable RHO/RAC effector that binds to the GTP-bound forms of RHO and RAC1. It probably binds p21 with a tighter specificity in vivo. Displays serine/threonine protein kinase activity. Plays an important role in the regulation of cytokinesis and the development of the central nervous system. Phosphorylates MYL9/MLC2.
Plays a role in cytokinesis. Required for KIF14 localization to the central spindle and midbody. Putative RHO/RAC effector that binds to the GTP-bound forms of RHO and RAC1. It probably binds p21 with a tighter specificity in vivo. Displays serine/threonine protein kinase activity. Plays an important role in the regulation of cytokinesis and the development of the central nervous system. Phosphorylates MYL9/MLC2.
Plays a role in cytokinesis. Required for KIF14 localization to the central spindle and midbody. Putative RHO/RAC effector that binds to the GTP-bound forms of RHO and RAC1. It probably binds p21 with a tighter specificity in vivo. Displays serine/threonine protein kinase activity. Plays an important role in the regulation of cytokinesis and the development of the central nervous system. Phosphorylates MYL9/MLC2.
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Subunit
Interacts with TTC3 (By similarity). Homodimer (Probable). Directly interacts with KIF14 depending on the activation state (stronger interaction with the kinase-dead form).
Directly interacts with KIF14 depending on the activation state (stronger interaction with the kinase-dead form). Homodimer (By similarity). Interacts with TTC3.
Directly interacts with KIF14 depending on the activation state (stronger interaction with the kinase-dead form). Homodimer (By similarity). Interacts with TTC3.
Similarity
Belongs to the protein kinase superfamily.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family. PKC subfamily.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family. PKC subfamily.
Keywords
Acetylation
Alternative splicing
ATP-binding
Cell cycle
Cell division
Coiled coil
Complete proteome
Cytoplasm
Developmental protein
Differentiation
Direct protein sequencing
Kinase
Metal-binding
Mitosis
Neurogenesis
Nucleotide-binding
Phosphoprotein
Reference proteome
SH3-binding
Transferase
Zinc
Zinc-finger
Disease mutation
Polymorphism
Primary microcephaly
Feature
chain Citron Rho-interacting kinase
splice variant In isoform 3.
sequence variant In dbSNP:rs36054900.
splice variant In isoform 3.
sequence variant In dbSNP:rs36054900.
Uniprot
A0A2H1VM13
A0A2A4JC55
A0A194PZP6
A0A212FML1
A0A2A4JCK2
A0A0N1PH16
+ More
H9JTJ4 A0A3S2M395 A0A087U0M7 A0A067QTR0 U4U1T4 A0A224YVE1 A7SF23 A0A131XYZ0 A0A1S3JLQ9 A0A139WGM0 A0A090X8E3 A0A1E1XP48 A0A3R7QQE6 A0A1I8PSJ6 K1QJU8 A0A2P2ID97 O88527 R7VBH7 A0A2R5LGF3 N6U8X4 A0A0M3QWL1 A0A182SLE6 P49025-2 A0A1I8NF21 W8BFU0 A0A1W4WKY3 A0A084W131 B4L0R1 A0A061I1R3 A0A061I4X5 A0A061I731 A0A1V9X907 A0A061I429 A0A0P6AEK9 Q2M0M0 G3GU69 A0A0N8D671 A0A0P5YXP0 A0A0P4VZM6 A0A164YA17 A0A091QEG4 A0A0P6HUK1 A0A2Y9SK60 A0A2J8MDX1 E9PSL7 A0A0L0CCY0 A0A0G2JWA9 B0XI46 O14578-2 B4GRG0 A0A3Q1N4A7 W5NR73 G3X146 W5NR76 A0A3Q1ML34 A0A3Q1M4D1 A0A2K5LKY4 A0A1B0EWQ0 A0A452FUE0 A0A452FUG8 A0A2P4SR47 A0A091WA01 E9HJU7 A0A2J8XK74 B3MB20 L8IJC4 A0A3B0KLI4 T2MJP6 E9QL53 B4N337 A0A182MUT5 F6SBR5 D3Z1U0 G1N9Y8 D3YU89 M3VVN3 F6PMM0 M3WV24 A0A210Q6D6 F6T1H9 A0A3Q2I696 A0A3Q2H051 F6PUR0 A0A3Q2HN58 A0A0A0ANW0 D2HCN3 G1MC31 A0A0P5WBY1 B4HG27 A0A2I0LZ20 A0A182U1X0 R7VNX2 A0A182RJK1 A0A0J9RUE6
H9JTJ4 A0A3S2M395 A0A087U0M7 A0A067QTR0 U4U1T4 A0A224YVE1 A7SF23 A0A131XYZ0 A0A1S3JLQ9 A0A139WGM0 A0A090X8E3 A0A1E1XP48 A0A3R7QQE6 A0A1I8PSJ6 K1QJU8 A0A2P2ID97 O88527 R7VBH7 A0A2R5LGF3 N6U8X4 A0A0M3QWL1 A0A182SLE6 P49025-2 A0A1I8NF21 W8BFU0 A0A1W4WKY3 A0A084W131 B4L0R1 A0A061I1R3 A0A061I4X5 A0A061I731 A0A1V9X907 A0A061I429 A0A0P6AEK9 Q2M0M0 G3GU69 A0A0N8D671 A0A0P5YXP0 A0A0P4VZM6 A0A164YA17 A0A091QEG4 A0A0P6HUK1 A0A2Y9SK60 A0A2J8MDX1 E9PSL7 A0A0L0CCY0 A0A0G2JWA9 B0XI46 O14578-2 B4GRG0 A0A3Q1N4A7 W5NR73 G3X146 W5NR76 A0A3Q1ML34 A0A3Q1M4D1 A0A2K5LKY4 A0A1B0EWQ0 A0A452FUE0 A0A452FUG8 A0A2P4SR47 A0A091WA01 E9HJU7 A0A2J8XK74 B3MB20 L8IJC4 A0A3B0KLI4 T2MJP6 E9QL53 B4N337 A0A182MUT5 F6SBR5 D3Z1U0 G1N9Y8 D3YU89 M3VVN3 F6PMM0 M3WV24 A0A210Q6D6 F6T1H9 A0A3Q2I696 A0A3Q2H051 F6PUR0 A0A3Q2HN58 A0A0A0ANW0 D2HCN3 G1MC31 A0A0P5WBY1 B4HG27 A0A2I0LZ20 A0A182U1X0 R7VNX2 A0A182RJK1 A0A0J9RUE6
EC Number
2.7.11.1
Pubmed
26354079
22118469
19121390
24845553
23537049
28797301
+ More
17615350 18362917 19820115 25970599 29209593 22992520 9697773 23254933 8543060 9792683 16141072 15489334 11086988 16452087 18034455 21183079 25315136 24495485 24438588 17994087 23929341 28327890 15632085 21804562 15057822 22673903 26108605 15983625 10231032 16541075 16431929 16236794 17488780 18669648 19413330 19690332 19608861 20068231 21269460 21457715 21406692 22814378 23186163 27453578 27453579 27503289 27519304 17344846 19393038 20809919 21709235 21292972 22751099 24065732 19468303 20838655 17975172 19892987 28812685 20010809 23371554 22936249
17615350 18362917 19820115 25970599 29209593 22992520 9697773 23254933 8543060 9792683 16141072 15489334 11086988 16452087 18034455 21183079 25315136 24495485 24438588 17994087 23929341 28327890 15632085 21804562 15057822 22673903 26108605 15983625 10231032 16541075 16431929 16236794 17488780 18669648 19413330 19690332 19608861 20068231 21269460 21457715 21406692 22814378 23186163 27453578 27453579 27503289 27519304 17344846 19393038 20809919 21709235 21292972 22751099 24065732 19468303 20838655 17975172 19892987 28812685 20010809 23371554 22936249
EMBL
ODYU01003294
SOQ41863.1
NWSH01002014
PCG69429.1
KQ459582
KPI98802.1
+ More
AGBW02007651 OWR54966.1 PCG69428.1 KQ460208 KPJ16621.1 BABH01003698 BABH01003699 BABH01003700 BABH01003701 BABH01003702 RSAL01000051 RVE50300.1 KK117600 KFM70916.1 KK853478 KDR07314.1 KB632003 ERL87819.1 GFPF01006788 MAA17934.1 DS469641 EDO37677.1 GEFM01004314 JAP71482.1 KQ971347 KYB26957.1 GBIH01002004 JAC92706.1 GFAA01002693 JAU00742.1 QCYY01001877 ROT74596.1 JH816753 EKC34043.1 IACF01006400 LAB71983.1 AF070065 AAC27932.1 AMQN01000719 KB295062 ELU13656.1 GGLE01004480 MBY08606.1 APGK01044205 KB741021 ENN75042.1 CP012525 ALC44339.1 U39904 AF086823 AF086824 AK145037 AF070066 BC023775 BC051165 GAMC01014414 JAB92141.1 ATLV01019208 KE525264 KFB43925.1 CH933809 EDW18138.1 KE676001 ERE74492.1 ERE74494.1 ERE74495.1 MNPL01018732 OQR70024.1 ERE74493.1 GDIP01030393 JAM73322.1 CH379069 EAL30910.1 JH000026 EGV93151.1 GDIP01064879 JAM38836.1 GDIP01053130 JAM50585.1 GDRN01100663 GDRN01100662 JAI58523.1 LRGB01000930 KZS15038.1 KK674753 KFQ07934.1 GDIQ01013295 JAN81442.1 NBAG03000260 PNI57719.1 AABR07036527 AABR07036528 AABR07036529 AABR07036530 AABR07036531 JRES01000577 KNC30067.1 DS233256 EDS28986.1 AY257469 AY209000 AY681966 AB023166 AC002563 AC079317 AC004813 CH479188 EDW40345.1 AMGL01039306 AMGL01039307 AMGL01039308 AEFK01072816 AEFK01072817 AEFK01072818 AEFK01072819 AEFK01072820 AEFK01072821 AEFK01072822 AJWK01006467 LWLT01000018 PPHD01027952 POI26580.1 KK735113 KFR12457.1 GL732664 EFX67999.1 NDHI03003364 PNJ82444.1 CH902618 EDV40286.1 JH881290 ELR55202.1 OUUW01000012 SPP87439.1 HAAD01005910 CDG72142.1 AC115795 AC162461 CH964062 EDW78776.2 AXCM01008027 AANG04002472 NEDP02004827 OWF44281.1 KL872580 KGL96209.1 GL192692 EFB19632.1 ACTA01027909 ACTA01035909 ACTA01043908 ACTA01051908 ACTA01059908 GDIP01088224 JAM15491.1 CH480815 EDW41273.1 AKCR02000059 PKK22678.1 KB375996 EMC78645.1 CM002912 KMY99247.1
AGBW02007651 OWR54966.1 PCG69428.1 KQ460208 KPJ16621.1 BABH01003698 BABH01003699 BABH01003700 BABH01003701 BABH01003702 RSAL01000051 RVE50300.1 KK117600 KFM70916.1 KK853478 KDR07314.1 KB632003 ERL87819.1 GFPF01006788 MAA17934.1 DS469641 EDO37677.1 GEFM01004314 JAP71482.1 KQ971347 KYB26957.1 GBIH01002004 JAC92706.1 GFAA01002693 JAU00742.1 QCYY01001877 ROT74596.1 JH816753 EKC34043.1 IACF01006400 LAB71983.1 AF070065 AAC27932.1 AMQN01000719 KB295062 ELU13656.1 GGLE01004480 MBY08606.1 APGK01044205 KB741021 ENN75042.1 CP012525 ALC44339.1 U39904 AF086823 AF086824 AK145037 AF070066 BC023775 BC051165 GAMC01014414 JAB92141.1 ATLV01019208 KE525264 KFB43925.1 CH933809 EDW18138.1 KE676001 ERE74492.1 ERE74494.1 ERE74495.1 MNPL01018732 OQR70024.1 ERE74493.1 GDIP01030393 JAM73322.1 CH379069 EAL30910.1 JH000026 EGV93151.1 GDIP01064879 JAM38836.1 GDIP01053130 JAM50585.1 GDRN01100663 GDRN01100662 JAI58523.1 LRGB01000930 KZS15038.1 KK674753 KFQ07934.1 GDIQ01013295 JAN81442.1 NBAG03000260 PNI57719.1 AABR07036527 AABR07036528 AABR07036529 AABR07036530 AABR07036531 JRES01000577 KNC30067.1 DS233256 EDS28986.1 AY257469 AY209000 AY681966 AB023166 AC002563 AC079317 AC004813 CH479188 EDW40345.1 AMGL01039306 AMGL01039307 AMGL01039308 AEFK01072816 AEFK01072817 AEFK01072818 AEFK01072819 AEFK01072820 AEFK01072821 AEFK01072822 AJWK01006467 LWLT01000018 PPHD01027952 POI26580.1 KK735113 KFR12457.1 GL732664 EFX67999.1 NDHI03003364 PNJ82444.1 CH902618 EDV40286.1 JH881290 ELR55202.1 OUUW01000012 SPP87439.1 HAAD01005910 CDG72142.1 AC115795 AC162461 CH964062 EDW78776.2 AXCM01008027 AANG04002472 NEDP02004827 OWF44281.1 KL872580 KGL96209.1 GL192692 EFB19632.1 ACTA01027909 ACTA01035909 ACTA01043908 ACTA01051908 ACTA01059908 GDIP01088224 JAM15491.1 CH480815 EDW41273.1 AKCR02000059 PKK22678.1 KB375996 EMC78645.1 CM002912 KMY99247.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000053240
UP000005204
UP000283053
+ More
UP000054359 UP000027135 UP000030742 UP000001593 UP000085678 UP000007266 UP000283509 UP000095300 UP000005408 UP000014760 UP000019118 UP000092553 UP000075901 UP000000589 UP000095301 UP000192223 UP000030765 UP000009192 UP000030759 UP000192247 UP000001819 UP000001075 UP000076858 UP000248484 UP000002494 UP000037069 UP000002320 UP000005640 UP000008744 UP000009136 UP000002356 UP000007648 UP000233060 UP000092461 UP000291000 UP000053605 UP000000305 UP000007801 UP000268350 UP000007798 UP000075883 UP000001645 UP000011712 UP000002281 UP000242188 UP000053858 UP000008912 UP000001292 UP000053872 UP000075902 UP000075900
UP000054359 UP000027135 UP000030742 UP000001593 UP000085678 UP000007266 UP000283509 UP000095300 UP000005408 UP000014760 UP000019118 UP000092553 UP000075901 UP000000589 UP000095301 UP000192223 UP000030765 UP000009192 UP000030759 UP000192247 UP000001819 UP000001075 UP000076858 UP000248484 UP000002494 UP000037069 UP000002320 UP000005640 UP000008744 UP000009136 UP000002356 UP000007648 UP000233060 UP000092461 UP000291000 UP000053605 UP000000305 UP000007801 UP000268350 UP000007798 UP000075883 UP000001645 UP000011712 UP000002281 UP000242188 UP000053858 UP000008912 UP000001292 UP000053872 UP000075902 UP000075900
Pfam
Interpro
IPR008271
Ser/Thr_kinase_AS
+ More
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR017405 Citron_Rho-interacting_kinase
IPR000719 Prot_kinase_dom
IPR000961 AGC-kinase_C
IPR029875 LET-502
IPR001180 CNH_dom
IPR017892 Pkinase_C
IPR037708 CRIK_dom
IPR001849 PH_domain
IPR002219 PE/DAG-bd
IPR011993 PH-like_dom_sf
IPR013604 7TM_chemorcpt
IPR007000 PLipase_B-like
IPR025768 TFG_box
IPR025761 FFD_box
IPR010920 LSM_dom_sf
IPR019050 FDF_dom
IPR025762 DFDF
IPR025609 Lsm14-like_N
IPR004152 GAT_dom
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR017405 Citron_Rho-interacting_kinase
IPR000719 Prot_kinase_dom
IPR000961 AGC-kinase_C
IPR029875 LET-502
IPR001180 CNH_dom
IPR017892 Pkinase_C
IPR037708 CRIK_dom
IPR001849 PH_domain
IPR002219 PE/DAG-bd
IPR011993 PH-like_dom_sf
IPR013604 7TM_chemorcpt
IPR007000 PLipase_B-like
IPR025768 TFG_box
IPR025761 FFD_box
IPR010920 LSM_dom_sf
IPR019050 FDF_dom
IPR025762 DFDF
IPR025609 Lsm14-like_N
IPR004152 GAT_dom
Gene 3D
ProteinModelPortal
A0A2H1VM13
A0A2A4JC55
A0A194PZP6
A0A212FML1
A0A2A4JCK2
A0A0N1PH16
+ More
H9JTJ4 A0A3S2M395 A0A087U0M7 A0A067QTR0 U4U1T4 A0A224YVE1 A7SF23 A0A131XYZ0 A0A1S3JLQ9 A0A139WGM0 A0A090X8E3 A0A1E1XP48 A0A3R7QQE6 A0A1I8PSJ6 K1QJU8 A0A2P2ID97 O88527 R7VBH7 A0A2R5LGF3 N6U8X4 A0A0M3QWL1 A0A182SLE6 P49025-2 A0A1I8NF21 W8BFU0 A0A1W4WKY3 A0A084W131 B4L0R1 A0A061I1R3 A0A061I4X5 A0A061I731 A0A1V9X907 A0A061I429 A0A0P6AEK9 Q2M0M0 G3GU69 A0A0N8D671 A0A0P5YXP0 A0A0P4VZM6 A0A164YA17 A0A091QEG4 A0A0P6HUK1 A0A2Y9SK60 A0A2J8MDX1 E9PSL7 A0A0L0CCY0 A0A0G2JWA9 B0XI46 O14578-2 B4GRG0 A0A3Q1N4A7 W5NR73 G3X146 W5NR76 A0A3Q1ML34 A0A3Q1M4D1 A0A2K5LKY4 A0A1B0EWQ0 A0A452FUE0 A0A452FUG8 A0A2P4SR47 A0A091WA01 E9HJU7 A0A2J8XK74 B3MB20 L8IJC4 A0A3B0KLI4 T2MJP6 E9QL53 B4N337 A0A182MUT5 F6SBR5 D3Z1U0 G1N9Y8 D3YU89 M3VVN3 F6PMM0 M3WV24 A0A210Q6D6 F6T1H9 A0A3Q2I696 A0A3Q2H051 F6PUR0 A0A3Q2HN58 A0A0A0ANW0 D2HCN3 G1MC31 A0A0P5WBY1 B4HG27 A0A2I0LZ20 A0A182U1X0 R7VNX2 A0A182RJK1 A0A0J9RUE6
H9JTJ4 A0A3S2M395 A0A087U0M7 A0A067QTR0 U4U1T4 A0A224YVE1 A7SF23 A0A131XYZ0 A0A1S3JLQ9 A0A139WGM0 A0A090X8E3 A0A1E1XP48 A0A3R7QQE6 A0A1I8PSJ6 K1QJU8 A0A2P2ID97 O88527 R7VBH7 A0A2R5LGF3 N6U8X4 A0A0M3QWL1 A0A182SLE6 P49025-2 A0A1I8NF21 W8BFU0 A0A1W4WKY3 A0A084W131 B4L0R1 A0A061I1R3 A0A061I4X5 A0A061I731 A0A1V9X907 A0A061I429 A0A0P6AEK9 Q2M0M0 G3GU69 A0A0N8D671 A0A0P5YXP0 A0A0P4VZM6 A0A164YA17 A0A091QEG4 A0A0P6HUK1 A0A2Y9SK60 A0A2J8MDX1 E9PSL7 A0A0L0CCY0 A0A0G2JWA9 B0XI46 O14578-2 B4GRG0 A0A3Q1N4A7 W5NR73 G3X146 W5NR76 A0A3Q1ML34 A0A3Q1M4D1 A0A2K5LKY4 A0A1B0EWQ0 A0A452FUE0 A0A452FUG8 A0A2P4SR47 A0A091WA01 E9HJU7 A0A2J8XK74 B3MB20 L8IJC4 A0A3B0KLI4 T2MJP6 E9QL53 B4N337 A0A182MUT5 F6SBR5 D3Z1U0 G1N9Y8 D3YU89 M3VVN3 F6PMM0 M3WV24 A0A210Q6D6 F6T1H9 A0A3Q2I696 A0A3Q2H051 F6PUR0 A0A3Q2HN58 A0A0A0ANW0 D2HCN3 G1MC31 A0A0P5WBY1 B4HG27 A0A2I0LZ20 A0A182U1X0 R7VNX2 A0A182RJK1 A0A0J9RUE6
PDB
4AW2
E-value=2.32345e-40,
Score=413
Ontologies
GO
GO:0005524
GO:0004672
GO:0004674
GO:0000281
GO:0035556
GO:0046872
GO:0005737
GO:0018107
GO:0005856
GO:0031032
GO:0004697
GO:0032154
GO:0050909
GO:0030866
GO:0016021
GO:0015629
GO:0045665
GO:0017124
GO:0030496
GO:0007030
GO:0043025
GO:0005829
GO:0030165
GO:0005773
GO:0001726
GO:0000086
GO:0008064
GO:0005634
GO:0016358
GO:0050774
GO:0007091
GO:0048699
GO:0097110
GO:0007283
GO:0005886
GO:0032467
GO:0000278
GO:0000070
GO:0031985
GO:0051402
GO:0006325
GO:2000431
GO:0000915
GO:0007112
GO:0005826
GO:0001889
GO:0016020
GO:0005623
GO:0006886
GO:0006468
GO:0006413
GO:0031369
GO:0003723
GO:0006730
GO:0009258
GO:0016155
GO:0003824
PANTHER
Topology
Subcellular location
Cytoplasm
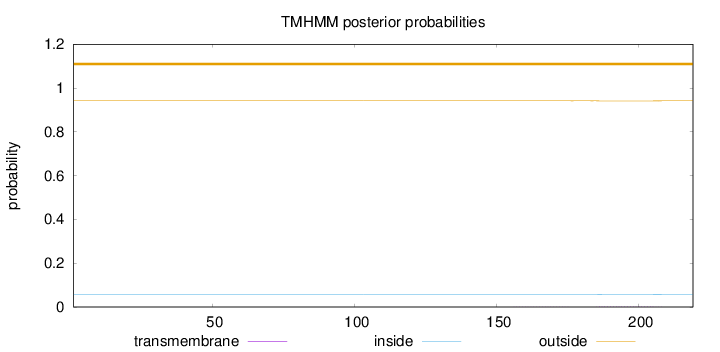
Length:
219
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0441
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05851
outside
1 - 219
Population Genetic Test Statistics
Pi
186.952941
Theta
177.955701
Tajima's D
0.339012
CLR
0.907844
CSRT
0.465576721163942
Interpretation
Uncertain
