Pre Gene Modal
BGIBMGA012855
Annotation
gephyrin_[Spodoptera_littoralis]
Full name
Molybdopterin molybdenumtransferase
+ More
Molybdenum cofactor synthesis protein cinnamon
Molybdenum cofactor synthesis protein cinnamon
Location in the cell
Cytoplasmic Reliability : 1.606
Sequence
CDS
ATGATAAAAGCAGTAGCAGTCATCACTGTGAGTGACACCTGTTTCAAAGATAATTCTAAAGATGAATCTGGTCCAGCACTTTGCCATTTAGTTAAAAAATTATTTCCTGACTCCAATTTACACACAATAATAGTACCTGATGAACGAGAAATGATTGAAAGGGAACTGAAATATTTTTGTGATAGTAATCTAGACTTAATTCTAACAACAGGAGGAACAGGTCTAAGCCCAAGGGATGTCACTCCAGAGGCTACAAAAGTTGTTATACACAAAGAAATTCCCAGTATATCTGTAGCCTTAACTCTGGAGAGCTTAAAAAAAACTCCTATGGCTATGCTTTCAAGATCTGTAGCAGGATTGCGTGATAAAACTTTAATCGTAAATTTACCAGGAAGCAAAAAAGCTGTCTTGGAATGCTTTGAAGTCATCAAACCAATTCTGCCTCATGCAATAGCATTAGTACGGAATGAATTGTCAGAAGTTAGAACAATACATGATTCATTGCAATTTAGTCATTTGTGTCCACATAGGAGCAACGTAGATATTACAAAGATAGCACTAAGACCAAGAGCATCCCCATATTCAATGATTGAAATGGCTGAAGCCTGTAAGATTGTAGATTCAGTAATGGGACAGTGGAATGAAATGTTAGAGATTGTACCTTTAGATCAAGCACTTGGTAGGGTAGTTGCTCAACCTCTGCGTGCAAAAGAGCCAATGCCGCCATTTCCAGCTTCCGTTAAAGATGGGTATGCGTGTCTGAGTACGGACGGTGTTGGGCCTAGAAAAGTGAATGCTGCTATTACAGCGGGTGATGCTCCGACTACACCGCTTTGTATGGGCGAGTGTGTGCGCATCAATACCGGCGCTGCATTACCTGCTGGTGCCGATTGCGTTGTACAAGTTGAGGATACAAAATTGCTATTAGCGTCTGACGATGGCGTGACTGAGTTGGAAATCGAGATACTGATCGCACCTGAGCCAGGTCAAGATGTTAGACCTGTTGGTTTTGATATAGCAATGGGGGCCACGCTTGTAGACAAAGGTGAATCTATAGATGCAGCTAAAATAGGAGTTTTAGCAGGAGCTGGTTACCAGTCGGTCACTGTTCGCGTCGGTCCCACGGTGGCGCTGATGTCAACTGGAAACGAATTACAAGAACCGTCCGAATTGAAACTCAGGCCAGCACACATTCGGGACTCCAATAGAGCGATGCTGAGCGCCTTGTTGAAAGAGCACGGGTACCAGAGTATAGATTGCGGTATTTGTCGTGACTCTCCCGAAGAGCTAGTGGCCGGTATATCGGCCGCCCTGACCAAGGCCGACGTGTTGGTCTGCACCGGAGGCGTGTCGATGGGCGAGAAAGATTTACTGAAGCCAGTATTATTAAACGATTTCGGTGCAACACTTCACTATGGACGCGTTCGCATGAAGCCCGGCAAGCCGTCGACTTTTGCAACATGCCAGTTTGAAGGAAAAACTAAATTTATTTTTGCATTGCCTGGTAATCCAGTTTCTGCCTACGTTTGCTGTTTGCTGTTCGTGATCCGAGGTCTGCGCGTGTGCAGCGGCAAGAGGGGCTCGTGGCCCAGACTCCGCGCCAAGCTCGCGCACGGCGTCGCGCTCGACCCCCGCCCGGAGTACGCGCGGGCCGACATCGACGTGTCCCAGGACGACGACATGCCCACTGTGAAGTTGTTGGGCAACCAGTGCAGTAGTCGTCTGTTGAGCGCGTGCGGTGCGAGCGTGCTGGTGGAGTTGCCGGCCGCCTCCCCCGCGCACCCCCGGCTGCCGGCCGGCGCCATCGTGCCCGCGCTCGTCACCGGGACGCTACACAACAGCGCAGTCTAA
Protein
MIKAVAVITVSDTCFKDNSKDESGPALCHLVKKLFPDSNLHTIIVPDEREMIERELKYFCDSNLDLILTTGGTGLSPRDVTPEATKVVIHKEIPSISVALTLESLKKTPMAMLSRSVAGLRDKTLIVNLPGSKKAVLECFEVIKPILPHAIALVRNELSEVRTIHDSLQFSHLCPHRSNVDITKIALRPRASPYSMIEMAEACKIVDSVMGQWNEMLEIVPLDQALGRVVAQPLRAKEPMPPFPASVKDGYACLSTDGVGPRKVNAAITAGDAPTTPLCMGECVRINTGAALPAGADCVVQVEDTKLLLASDDGVTELEIEILIAPEPGQDVRPVGFDIAMGATLVDKGESIDAAKIGVLAGAGYQSVTVRVGPTVALMSTGNELQEPSELKLRPAHIRDSNRAMLSALLKEHGYQSIDCGICRDSPEELVAGISAALTKADVLVCTGGVSMGEKDLLKPVLLNDFGATLHYGRVRMKPGKPSTFATCQFEGKTKFIFALPGNPVSAYVCCLLFVIRGLRVCSGKRGSWPRLRAKLAHGVALDPRPEYARADIDVSQDDDMPTVKLLGNQCSSRLLSACGASVLVELPAASPAHPRLPAGAIVPALVTGTLHNSAV
Summary
Description
Catalyzes two steps in the biosynthesis of the molybdenum cofactor. In the first step, molybdopterin is adenylated. Subsequently, molybdate is inserted into adenylated molybdopterin and AMP is released.
Catalytic Activity
ATP + H(+) + molybdopterin = adenylyl-molybdopterin + diphosphate
adenylyl-molybdopterin + H(+) + molybdate = AMP + H2O + Mo-molybdopterin
adenylyl-molybdopterin + H(+) + molybdate = AMP + H2O + Mo-molybdopterin
Cofactor
Mg(2+)
Similarity
Belongs to the MoeA family.
Belongs to the heat shock protein 70 family.
In the N-terminal section; belongs to the MoaB/Mog family.
In the C-terminal section; belongs to the MoeA family.
Belongs to the heat shock protein 70 family.
In the N-terminal section; belongs to the MoaB/Mog family.
In the C-terminal section; belongs to the MoeA family.
Keywords
ATP-binding
Complete proteome
Magnesium
Metal-binding
Molybdenum
Molybdenum cofactor biosynthesis
Multifunctional enzyme
Nucleotide-binding
Phosphoprotein
Reference proteome
Transferase
Feature
chain Molybdopterin molybdenumtransferase
Uniprot
H9JTJ3
A0A2Z6BY24
A0A2H1VM10
A0A0U2LXC6
A0A2A4JBH6
A0A212FMQ0
+ More
A0A194Q1D2 A0A3M6T7T7 A0A1S3IKU0 A0A1E1X3I7 A0A2R5LID8 A0A154PNI4 A0A1Y1KVI4 A0A067RL79 A0A158NG88 A0A0C9RJR3 A0A369S7X3 L7ME56 B3SAE9 A7SKS5 A0A224YTW3 A0A3L8DFD2 A0A1Z5KY69 A0A023F1L8 A0A2B4SD59 T1JB58 A0A232F6C1 A0A069DVR5 A0A2A3E8R6 A0A3R7SWB1 A0A0V0G894 A0A164RYU0 D2A001 E9GEV6 A0A0P6AXP2 A0A0N8B2E5 A0A1B6L8C5 A0A0P5U1T8 A0A0L7QWC3 E2CA55 A0A026W058 A0A0P5CU31 A0A195B4G7 E2AD81 R7TMH7 A0A151X1Q5 A0A0M9ABH7 F4WZE8 K7J7P8 A0A1B6DQQ3 A0A443RR44 A0A0P6HI74 A0A1D2M7V6 A0A195E553 A0A195ET58 A0A146M710 A0A226EQA4 T1HE94 A0A0J7KLE8 A0A1B6CEV1 A0A2G8K2K3 A0A1D1Z389 A0A2G8K1H2 B4I901 A0A1D1VH67 A0A132A7D9 A0A444SJJ1 A0A1B6D375 B4NCY1 A0A1W4UVP0 B3P948 P39205 B4PWJ6 A0A2Z6RGY1 A0A0L0C517 E4Y6H7 G3MGC9 A0A397J9P6 E4XI79 A0A2I1G4K1 A0A015KN09 U9SKS5 A0A2N0PVF3 A0A2I1EHL4 A0A1I8MLI6 A0A372R2G5 A0A443S6R7 A0A367KYJ7 A0A397SZR3 A0A2N1NXK8 A0A3M7QVB2 W5JCU1 A0A0V0VK02 A0A0C7AZE5 W8AH29 A0A0V1CXH5 A0A0V1LSY7 A0A0V0Z751 A0A077X3V0
A0A194Q1D2 A0A3M6T7T7 A0A1S3IKU0 A0A1E1X3I7 A0A2R5LID8 A0A154PNI4 A0A1Y1KVI4 A0A067RL79 A0A158NG88 A0A0C9RJR3 A0A369S7X3 L7ME56 B3SAE9 A7SKS5 A0A224YTW3 A0A3L8DFD2 A0A1Z5KY69 A0A023F1L8 A0A2B4SD59 T1JB58 A0A232F6C1 A0A069DVR5 A0A2A3E8R6 A0A3R7SWB1 A0A0V0G894 A0A164RYU0 D2A001 E9GEV6 A0A0P6AXP2 A0A0N8B2E5 A0A1B6L8C5 A0A0P5U1T8 A0A0L7QWC3 E2CA55 A0A026W058 A0A0P5CU31 A0A195B4G7 E2AD81 R7TMH7 A0A151X1Q5 A0A0M9ABH7 F4WZE8 K7J7P8 A0A1B6DQQ3 A0A443RR44 A0A0P6HI74 A0A1D2M7V6 A0A195E553 A0A195ET58 A0A146M710 A0A226EQA4 T1HE94 A0A0J7KLE8 A0A1B6CEV1 A0A2G8K2K3 A0A1D1Z389 A0A2G8K1H2 B4I901 A0A1D1VH67 A0A132A7D9 A0A444SJJ1 A0A1B6D375 B4NCY1 A0A1W4UVP0 B3P948 P39205 B4PWJ6 A0A2Z6RGY1 A0A0L0C517 E4Y6H7 G3MGC9 A0A397J9P6 E4XI79 A0A2I1G4K1 A0A015KN09 U9SKS5 A0A2N0PVF3 A0A2I1EHL4 A0A1I8MLI6 A0A372R2G5 A0A443S6R7 A0A367KYJ7 A0A397SZR3 A0A2N1NXK8 A0A3M7QVB2 W5JCU1 A0A0V0VK02 A0A0C7AZE5 W8AH29 A0A0V1CXH5 A0A0V1LSY7 A0A0V0Z751 A0A077X3V0
EC Number
2.10.1.1
Pubmed
19121390
22118469
26354079
30382153
28503490
28004739
+ More
24845553 21347285 30042472 25576852 18719581 17615350 28797301 30249741 28528879 25474469 28648823 26334808 18362917 19820115 21292972 20798317 24508170 23254933 21719571 20075255 27289101 26823975 29023486 17994087 27649274 26555130 8088525 10731132 10394904 12537572 10731137 12537569 17372656 17550304 26108605 21097902 22216098 24277808 30271968 29355972 25315136 29674435 30375419 20920257 23761445 24495485
24845553 21347285 30042472 25576852 18719581 17615350 28797301 30249741 28528879 25474469 28648823 26334808 18362917 19820115 21292972 20798317 24508170 23254933 21719571 20075255 27289101 26823975 29023486 17994087 27649274 26555130 8088525 10731132 10394904 12537572 10731137 12537569 17372656 17550304 26108605 21097902 22216098 24277808 30271968 29355972 25315136 29674435 30375419 20920257 23761445 24495485
EMBL
BABH01003706
LC375276
LC375277
BBD34849.1
BBD34850.1
ODYU01003294
+ More
SOQ41860.1 KP308211 ALH43429.1 NWSH01002014 PCG69425.1 AGBW02007651 OWR54969.1 KQ459582 KPI98799.1 RCHS01004153 RMX37354.1 GFAC01005363 JAT93825.1 GGLE01005059 MBY09185.1 KQ435005 KZC13403.1 GEZM01078176 GEZM01078175 JAV62887.1 KK852559 KDR21365.1 ADTU01001578 GBYB01002943 GBYB01008440 JAG72710.1 JAG78207.1 NOWV01000053 RDD42544.1 GACK01002947 JAA62087.1 DS985261 EDV20302.1 DS469691 EDO35668.1 GFPF01009902 MAA21048.1 QOIP01000009 RLU19094.1 GFJQ02006975 JAV99994.1 GBBI01003823 JAC14889.1 LSMT01000128 PFX26375.1 JH432008 NNAY01000808 OXU26406.1 GBGD01000889 JAC88000.1 KZ288323 PBC28115.1 QCYY01001399 ROT78327.1 GECL01001820 JAP04304.1 LRGB01002121 KZS09070.1 KQ971338 EFA02449.2 GL732541 EFX82014.1 GDIP01029548 JAM74167.1 GDIQ01219499 JAK32226.1 GEBQ01020020 JAT19957.1 GDIP01118981 JAL84733.1 KQ414716 KOC62861.1 GL453920 EFN75210.1 KK107519 EZA49402.1 GDIP01181674 JAJ41728.1 KQ976625 KYM79079.1 GL438685 EFN68596.1 AMQN01011973 KB309218 ELT95068.1 KQ982585 KYQ54343.1 KQ435692 KOX81080.1 GL888479 EGI60392.1 AAZX01003037 GEDC01009365 JAS27933.1 NCKU01000040 RWS17754.1 GDIQ01027752 JAN66985.1 LJIJ01002957 ODM89039.1 KQ979609 KYN20216.1 KQ981979 KYN31418.1 GDHC01003554 JAQ15075.1 LNIX01000002 OXA59805.1 ACPB03013340 LBMM01005921 KMQ91092.1 GEDC01025336 GEDC01002885 JAS11962.1 JAS34413.1 MRZV01000950 PIK42231.1 GDJX01006563 JAT61373.1 MRZV01000983 PIK41858.1 CH480825 EDW43682.1 BDGG01000006 GAV00972.1 JXLN01011121 KPM06799.1 SAUD01034078 RXG53584.1 GEDC01017146 JAS20152.1 CH964239 EDW82690.1 CH954183 EDV45344.1 L19876 AE014298 AL050231 AY069078 CM000162 EDX00765.1 BEXD01001013 GBB91728.1 JRES01000981 KNC26529.1 FN654296 CBY31227.1 JO840930 AEO32547.1 PQFF01000097 RHZ82706.1 FN653054 CBY10280.1 LLXI01000155 PKY41567.1 JEMT01025808 EXX61116.1 KI301032 BDIQ01000011 AUPC02000026 ERZ95706.1 GBC12727.1 POG79240.1 LLXJ01000353 PKC10821.1 LLXH01000459 LLXK01000449 PKC66500.1 PKY21607.1 QKKE01000213 RGB34170.1 NCKV01006869 RWS23230.1 PJQM01000007 RCI07283.1 QKYT01000218 RIA89485.1 LLXL01000076 PKK78603.1 REGN01005052 RNA15041.1 ADMH02001876 ETN60700.1 JYDN01000032 KRX63363.1 CCYT01000002 CEG64425.1 GAMC01018790 JAB87765.1 JYDI01000077 KRY53927.1 JYDW01000009 KRZ62471.1 JYDQ01000343 KRY08326.1 LK023386 CDS14290.1
SOQ41860.1 KP308211 ALH43429.1 NWSH01002014 PCG69425.1 AGBW02007651 OWR54969.1 KQ459582 KPI98799.1 RCHS01004153 RMX37354.1 GFAC01005363 JAT93825.1 GGLE01005059 MBY09185.1 KQ435005 KZC13403.1 GEZM01078176 GEZM01078175 JAV62887.1 KK852559 KDR21365.1 ADTU01001578 GBYB01002943 GBYB01008440 JAG72710.1 JAG78207.1 NOWV01000053 RDD42544.1 GACK01002947 JAA62087.1 DS985261 EDV20302.1 DS469691 EDO35668.1 GFPF01009902 MAA21048.1 QOIP01000009 RLU19094.1 GFJQ02006975 JAV99994.1 GBBI01003823 JAC14889.1 LSMT01000128 PFX26375.1 JH432008 NNAY01000808 OXU26406.1 GBGD01000889 JAC88000.1 KZ288323 PBC28115.1 QCYY01001399 ROT78327.1 GECL01001820 JAP04304.1 LRGB01002121 KZS09070.1 KQ971338 EFA02449.2 GL732541 EFX82014.1 GDIP01029548 JAM74167.1 GDIQ01219499 JAK32226.1 GEBQ01020020 JAT19957.1 GDIP01118981 JAL84733.1 KQ414716 KOC62861.1 GL453920 EFN75210.1 KK107519 EZA49402.1 GDIP01181674 JAJ41728.1 KQ976625 KYM79079.1 GL438685 EFN68596.1 AMQN01011973 KB309218 ELT95068.1 KQ982585 KYQ54343.1 KQ435692 KOX81080.1 GL888479 EGI60392.1 AAZX01003037 GEDC01009365 JAS27933.1 NCKU01000040 RWS17754.1 GDIQ01027752 JAN66985.1 LJIJ01002957 ODM89039.1 KQ979609 KYN20216.1 KQ981979 KYN31418.1 GDHC01003554 JAQ15075.1 LNIX01000002 OXA59805.1 ACPB03013340 LBMM01005921 KMQ91092.1 GEDC01025336 GEDC01002885 JAS11962.1 JAS34413.1 MRZV01000950 PIK42231.1 GDJX01006563 JAT61373.1 MRZV01000983 PIK41858.1 CH480825 EDW43682.1 BDGG01000006 GAV00972.1 JXLN01011121 KPM06799.1 SAUD01034078 RXG53584.1 GEDC01017146 JAS20152.1 CH964239 EDW82690.1 CH954183 EDV45344.1 L19876 AE014298 AL050231 AY069078 CM000162 EDX00765.1 BEXD01001013 GBB91728.1 JRES01000981 KNC26529.1 FN654296 CBY31227.1 JO840930 AEO32547.1 PQFF01000097 RHZ82706.1 FN653054 CBY10280.1 LLXI01000155 PKY41567.1 JEMT01025808 EXX61116.1 KI301032 BDIQ01000011 AUPC02000026 ERZ95706.1 GBC12727.1 POG79240.1 LLXJ01000353 PKC10821.1 LLXH01000459 LLXK01000449 PKC66500.1 PKY21607.1 QKKE01000213 RGB34170.1 NCKV01006869 RWS23230.1 PJQM01000007 RCI07283.1 QKYT01000218 RIA89485.1 LLXL01000076 PKK78603.1 REGN01005052 RNA15041.1 ADMH02001876 ETN60700.1 JYDN01000032 KRX63363.1 CCYT01000002 CEG64425.1 GAMC01018790 JAB87765.1 JYDI01000077 KRY53927.1 JYDW01000009 KRZ62471.1 JYDQ01000343 KRY08326.1 LK023386 CDS14290.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000275408
UP000085678
+ More
UP000076502 UP000027135 UP000005205 UP000253843 UP000009022 UP000001593 UP000279307 UP000225706 UP000215335 UP000242457 UP000283509 UP000076858 UP000007266 UP000000305 UP000053825 UP000008237 UP000053097 UP000078540 UP000000311 UP000014760 UP000075809 UP000053105 UP000007755 UP000002358 UP000285301 UP000094527 UP000078492 UP000078541 UP000198287 UP000015103 UP000036403 UP000230750 UP000001292 UP000186922 UP000288706 UP000007798 UP000192221 UP000008711 UP000000803 UP000002282 UP000247702 UP000037069 UP000266861 UP000234323 UP000022910 UP000018888 UP000236242 UP000232722 UP000232688 UP000095301 UP000263633 UP000288716 UP000253551 UP000265703 UP000233469 UP000276133 UP000000673 UP000054681 UP000054919 UP000054653 UP000054721 UP000054783
UP000076502 UP000027135 UP000005205 UP000253843 UP000009022 UP000001593 UP000279307 UP000225706 UP000215335 UP000242457 UP000283509 UP000076858 UP000007266 UP000000305 UP000053825 UP000008237 UP000053097 UP000078540 UP000000311 UP000014760 UP000075809 UP000053105 UP000007755 UP000002358 UP000285301 UP000094527 UP000078492 UP000078541 UP000198287 UP000015103 UP000036403 UP000230750 UP000001292 UP000186922 UP000288706 UP000007798 UP000192221 UP000008711 UP000000803 UP000002282 UP000247702 UP000037069 UP000266861 UP000234323 UP000022910 UP000018888 UP000236242 UP000232722 UP000232688 UP000095301 UP000263633 UP000288716 UP000253551 UP000265703 UP000233469 UP000276133 UP000000673 UP000054681 UP000054919 UP000054653 UP000054721 UP000054783
Pfam
Interpro
IPR041679
DNA2/NAM7-like_AAA
+ More
IPR005111 MoeA_C_domain_IV
IPR008284 MoCF_biosynth_CS
IPR001453 MoaB/Mog_dom
IPR036135 MoeA_linker/N_sf
IPR014808 DNA_replication_fac_Dna2_N
IPR027417 P-loop_NTPase
IPR041677 DNA2/NAM7_AAA_11
IPR036688 MoeA_C_domain_IV_sf
IPR005110 MoeA_linker/N
IPR036425 MoaB/Mog-like_dom_sf
IPR038987 MoeA-like
IPR013126 Hsp_70_fam
IPR018181 Heat_shock_70_CS
IPR040397 SWAP
IPR019147 SWAP_N_domain
IPR024639 DNA_pol_e_bsu_N
IPR007185 DNA_pol_a/d/e_bsu
IPR005111 MoeA_C_domain_IV
IPR008284 MoCF_biosynth_CS
IPR001453 MoaB/Mog_dom
IPR036135 MoeA_linker/N_sf
IPR014808 DNA_replication_fac_Dna2_N
IPR027417 P-loop_NTPase
IPR041677 DNA2/NAM7_AAA_11
IPR036688 MoeA_C_domain_IV_sf
IPR005110 MoeA_linker/N
IPR036425 MoaB/Mog-like_dom_sf
IPR038987 MoeA-like
IPR013126 Hsp_70_fam
IPR018181 Heat_shock_70_CS
IPR040397 SWAP
IPR019147 SWAP_N_domain
IPR024639 DNA_pol_e_bsu_N
IPR007185 DNA_pol_a/d/e_bsu
Gene 3D
ProteinModelPortal
H9JTJ3
A0A2Z6BY24
A0A2H1VM10
A0A0U2LXC6
A0A2A4JBH6
A0A212FMQ0
+ More
A0A194Q1D2 A0A3M6T7T7 A0A1S3IKU0 A0A1E1X3I7 A0A2R5LID8 A0A154PNI4 A0A1Y1KVI4 A0A067RL79 A0A158NG88 A0A0C9RJR3 A0A369S7X3 L7ME56 B3SAE9 A7SKS5 A0A224YTW3 A0A3L8DFD2 A0A1Z5KY69 A0A023F1L8 A0A2B4SD59 T1JB58 A0A232F6C1 A0A069DVR5 A0A2A3E8R6 A0A3R7SWB1 A0A0V0G894 A0A164RYU0 D2A001 E9GEV6 A0A0P6AXP2 A0A0N8B2E5 A0A1B6L8C5 A0A0P5U1T8 A0A0L7QWC3 E2CA55 A0A026W058 A0A0P5CU31 A0A195B4G7 E2AD81 R7TMH7 A0A151X1Q5 A0A0M9ABH7 F4WZE8 K7J7P8 A0A1B6DQQ3 A0A443RR44 A0A0P6HI74 A0A1D2M7V6 A0A195E553 A0A195ET58 A0A146M710 A0A226EQA4 T1HE94 A0A0J7KLE8 A0A1B6CEV1 A0A2G8K2K3 A0A1D1Z389 A0A2G8K1H2 B4I901 A0A1D1VH67 A0A132A7D9 A0A444SJJ1 A0A1B6D375 B4NCY1 A0A1W4UVP0 B3P948 P39205 B4PWJ6 A0A2Z6RGY1 A0A0L0C517 E4Y6H7 G3MGC9 A0A397J9P6 E4XI79 A0A2I1G4K1 A0A015KN09 U9SKS5 A0A2N0PVF3 A0A2I1EHL4 A0A1I8MLI6 A0A372R2G5 A0A443S6R7 A0A367KYJ7 A0A397SZR3 A0A2N1NXK8 A0A3M7QVB2 W5JCU1 A0A0V0VK02 A0A0C7AZE5 W8AH29 A0A0V1CXH5 A0A0V1LSY7 A0A0V0Z751 A0A077X3V0
A0A194Q1D2 A0A3M6T7T7 A0A1S3IKU0 A0A1E1X3I7 A0A2R5LID8 A0A154PNI4 A0A1Y1KVI4 A0A067RL79 A0A158NG88 A0A0C9RJR3 A0A369S7X3 L7ME56 B3SAE9 A7SKS5 A0A224YTW3 A0A3L8DFD2 A0A1Z5KY69 A0A023F1L8 A0A2B4SD59 T1JB58 A0A232F6C1 A0A069DVR5 A0A2A3E8R6 A0A3R7SWB1 A0A0V0G894 A0A164RYU0 D2A001 E9GEV6 A0A0P6AXP2 A0A0N8B2E5 A0A1B6L8C5 A0A0P5U1T8 A0A0L7QWC3 E2CA55 A0A026W058 A0A0P5CU31 A0A195B4G7 E2AD81 R7TMH7 A0A151X1Q5 A0A0M9ABH7 F4WZE8 K7J7P8 A0A1B6DQQ3 A0A443RR44 A0A0P6HI74 A0A1D2M7V6 A0A195E553 A0A195ET58 A0A146M710 A0A226EQA4 T1HE94 A0A0J7KLE8 A0A1B6CEV1 A0A2G8K2K3 A0A1D1Z389 A0A2G8K1H2 B4I901 A0A1D1VH67 A0A132A7D9 A0A444SJJ1 A0A1B6D375 B4NCY1 A0A1W4UVP0 B3P948 P39205 B4PWJ6 A0A2Z6RGY1 A0A0L0C517 E4Y6H7 G3MGC9 A0A397J9P6 E4XI79 A0A2I1G4K1 A0A015KN09 U9SKS5 A0A2N0PVF3 A0A2I1EHL4 A0A1I8MLI6 A0A372R2G5 A0A443S6R7 A0A367KYJ7 A0A397SZR3 A0A2N1NXK8 A0A3M7QVB2 W5JCU1 A0A0V0VK02 A0A0C7AZE5 W8AH29 A0A0V1CXH5 A0A0V1LSY7 A0A0V0Z751 A0A077X3V0
PDB
1T3E
E-value=1.08241e-90,
Score=852
Ontologies
PATHWAY
GO
GO:0032324
GO:0006777
GO:0061598
GO:0005524
GO:0061599
GO:0018315
GO:0046872
GO:0005829
GO:0005737
GO:0016021
GO:0098970
GO:0072579
GO:0099634
GO:0007529
GO:0097112
GO:0099572
GO:0030425
GO:0045211
GO:0006260
GO:0003677
GO:0003887
GO:0002121
GO:0006413
GO:0031369
GO:0003723
GO:0006730
GO:0009258
GO:0016155
GO:0003824
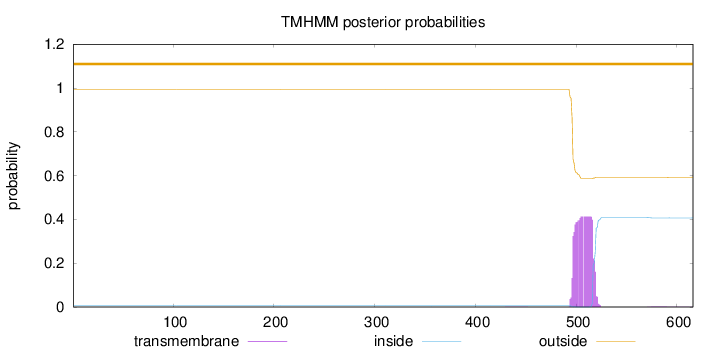
Topology
Length:
616
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.94226
Exp number, first 60 AAs:
3e-05
Total prob of N-in:
0.00542
outside
1 - 616
Population Genetic Test Statistics
Pi
244.212334
Theta
195.424762
Tajima's D
0.832416
CLR
0.156963
CSRT
0.611319434028299
Interpretation
Uncertain
