Pre Gene Modal
BGIBMGA012855
Annotation
PREDICTED:_DNA_replication_ATP-dependent_helicase/nuclease_DNA2_[Amyelois_transitella]
Full name
Ubiquinone biosynthesis O-methyltransferase, mitochondrial
+ More
DNA replication ATP-dependent helicase/nuclease DNA2
DNA replication ATP-dependent helicase/nuclease DNA2
Alternative Name
3-demethylubiquinol 3-O-methyltransferase
Polyprenyldihydroxybenzoate methyltransferase
DNA replication ATP-dependent helicase-like homolog
Polyprenyldihydroxybenzoate methyltransferase
DNA replication ATP-dependent helicase-like homolog
Location in the cell
Nuclear Reliability : 3.119
Sequence
CDS
ATGAATAAAGAAACAGATACGAATAAAAAATTAAAGCAAATTAAAATAGAAAATAAAGATCCTTTAAAACAACTTGAACATTCTCCAATATTCAACAGCTTACCTGATATATCAAATTTACAATGTTTCACATTTGACAAAAAAGATTTTGAACAAGATGATGTAAGTGATGTTTTCAATGATGATTGGGAGCACGAAGAGGATGCAGCAGCCTTTGACAAGTTAAATCTGAATAAAATGCAGAGATGTGAGGTAATCAGTTTGAAAAATCATGAAAACAAAATGGAACTTTTTTTAAAGAGCGGTGAAATGACTGGCACTTGTTTTGTTGAAGGGATATGGAAATCAATACCTTTGAGTCCGGGCGATATAATCAGTGTGATTGCAGCTAAAAATGAAACGGGGCAACATTATTTAAACGACACAACTGGGCTGCTCGTTCTCCGTCCGGACCACTTGATATCGAGCACAAGTGTTGTGGCTGGAGTGTTTTGTAAGCGAAAAGCTGTACTCCAAGAGAGATGGAGAGGAATAGACTCTGCCAATACGGCGATGACAATTGGAATAATGGTACATGAGCTAGTACAAAAGGCTCTGACACAACAAATATCAGATATAACTCAGTTACGTTTAGAAGCTGATAGAATGATAAGGGAGTCCATACAGATGTTGTACGACGCAGGGATGACCGAGGAGGAGACCCGGACAAGCATGCAGTGCTACATACAGCCTCTCAATGATTTCATGAAGAGCTATGTCGTTTCCAAGGCCAGTCATGGAATTATGCAAAATAACAAAGAAAACTGGAACGGTCATATTGATAAGGTTTTGGATATAGAAGAAAATATATGCTGTCCACAATTAGGACTGAAAGGGAAGATCGATGCCACTTTGCAAGTTACCGTACCTGAAAAAAATGGTCTCCAGAAAGCCATAGTTCCTTTAGAGTTGAAAAGTGGAAAGGCATCTGTGTCGGCAGAACATCGCGGCCAGCTCGTAATGTATGGAATGATGCTGAGCTTACACAGAAACGAAGACCCCACTGTGGCTTTACAACGTGGACTGTTGCTTTATTTGAGAGACAAAGTAGACGTGAGGGAGGTTAACTGTGGATACCCAGAGAGGCGCGACTTGGTAATGCTGAGGAATCAGTTAGTTGAAAATCTCGCTTCAGATCCCAAAGATCCAGATCCAGAACAGTTAACTGATATTGAAGATGCCTCATTTTTACTTCAGCAAAAGCTACCAGAACCGATCAATCATGAGAGAGCATGCACTAAATGTCCGTACTTATCCATTTGTTCTGTCCATTTGTGGCATTCGGGAGGCCCATCAGTCTCTGAAAATCATCCGCTGTCAAAGTTACAAGGCCAAGCACTAGGACACTTATCCAAAGGTCACATAAAATATTTCTTTCATTGGACGGCTTTACTTAAGATGGAAGAGAAATTTCAAACGAATTCATCACCTTTACAAGCCTTGTGGACTGAGAGTGCTGAAAAAAGAGAAAAAAGAGGGACTTGCATTCCCAATCTGAAAATAAAGGCAGTGGAGCAATCAGGTGAAAAACATCTTCACGTATTTCGACGTTCAGGTGTTAGATTAAATAGAGAGAAAGGACCTCAAGAGGGGGAGTTCGCAATAGTCAGTATCGAGGGCAGACCGTGGGTAGCCGCGGGAGTTGTCAGCTCAGTTAGTAGCGATGAAGTGCAGATACATTTGGAAAGAGACTTGAGTAAACGTCTCTGCGAAGACACATTGTACCACATCGATTCGTATGAGTCTTATTCAACTATTGTACAAAATCTGTCCAACCTCGGCGTCTTGCTTGAATACAGTGAAAGGTCGGAGCGGCTTCGAACTTTGATAATAGACAAAACGAAACCTCAGTTTGAGGCGAAGCTACCACGCGAAGTCGGCCGGCTCGGCATCAAGCTCATGCGCTCACTGAACATCGAACAGCAGCGAGCTGTACTAAAAGCTCTTTCTGCCCAGGATTACGCACTCCTCCAAGGTCTCCCGGGAACTGGGAAAACTCAAACCATTTCCGTGCTGATACAAATGCTAGTTGGACTCAAACAAAGAGTACTAGTGACTGCTCACACGCACTCGGCGGTCGACACTCTGTTGAGTAGATTACCAGAGACTATGAAAGTGATGCGGTTGGGTTCGACGGCACGTGTTGATATAAATCTGCTACATCGCACCGAGAAACAATTAACAGCTTCTTGTCGCACCGTCGAAGAGCTAGCAAGACTGTACGATTCAATGGAAGTCATTGGTGTAACATGTTTGGGGTCCACGCACGCCATACTCTCTAGAACGACGTTCGACATGTGCATTGTTGATGAAGCTACCCAGGTATTACAATGCACGGTATTGAGACCACTGTTTGCTGCTAGAAGATTCTTGCTAGTTGGTGATCCCAAACAACTCCCGCCTATCGTGAAAAGTCGCGCCGCAAGGCGTCTCGGCATGGAAGAGAGTCTGTTCCACAGATTAATGGACGAAGAAGCAACTTGCACCTTGCATCTGCAGTATCGCATGAATCAAGCTATAACCGATGTAGCGAATCAAGTAGCTTATGACTCGCGTTTGAAATGTGCCAACGGACAAGTTGCTCTAGCTACACTCAGTATTGATATGCAGAAATTATCATCAAAAACGGAATGGTTAAAGAGAGCTTGCAGTCCAGAACCTGAAGACGCTGTATTGTTTTTGGACATGAAATCTACAAATGTGTTTGATTTTGAAAATGAAAGCCGCTCGTGTAGGAACAAAGCCGAAGCTTGCCTAGTTCTCGACTTGATGGAATGCTTCAAAGAGGGTGGCGTTTCATCTAAAAATATAGGTATAATAGCGCCATATCGCGAACAAGTGTCACTACTGCGCAGGGCCCTGGCGTCCTACTCTGTAGAAGTTAACACTGTTGATCAGTTCCAAGGCAGAGACAAGTCCATTTTAATATATTCGTGTACCAAATGGAATGTAAGCGAAGATCATAAAGTTAAGGAAGGAGAGATACTGAACGACGATCGGAGATTAGCTGTGAGCGTGACTCGAGCGAAGCACAAGTTCATTATAGTTGGTAATTCAAAAGCGTTGCATTGCTACGTCCCACTGCAACGGCTGATACGAGCAAGCGCTACTGTCAGTCTCAATGATGATATTGTTAAATGTTTGTATCATAAGTATAAAGAAATCATGATTTAG
Protein
MNKETDTNKKLKQIKIENKDPLKQLEHSPIFNSLPDISNLQCFTFDKKDFEQDDVSDVFNDDWEHEEDAAAFDKLNLNKMQRCEVISLKNHENKMELFLKSGEMTGTCFVEGIWKSIPLSPGDIISVIAAKNETGQHYLNDTTGLLVLRPDHLISSTSVVAGVFCKRKAVLQERWRGIDSANTAMTIGIMVHELVQKALTQQISDITQLRLEADRMIRESIQMLYDAGMTEEETRTSMQCYIQPLNDFMKSYVVSKASHGIMQNNKENWNGHIDKVLDIEENICCPQLGLKGKIDATLQVTVPEKNGLQKAIVPLELKSGKASVSAEHRGQLVMYGMMLSLHRNEDPTVALQRGLLLYLRDKVDVREVNCGYPERRDLVMLRNQLVENLASDPKDPDPEQLTDIEDASFLLQQKLPEPINHERACTKCPYLSICSVHLWHSGGPSVSENHPLSKLQGQALGHLSKGHIKYFFHWTALLKMEEKFQTNSSPLQALWTESAEKREKRGTCIPNLKIKAVEQSGEKHLHVFRRSGVRLNREKGPQEGEFAIVSIEGRPWVAAGVVSSVSSDEVQIHLERDLSKRLCEDTLYHIDSYESYSTIVQNLSNLGVLLEYSERSERLRTLIIDKTKPQFEAKLPREVGRLGIKLMRSLNIEQQRAVLKALSAQDYALLQGLPGTGKTQTISVLIQMLVGLKQRVLVTAHTHSAVDTLLSRLPETMKVMRLGSTARVDINLLHRTEKQLTASCRTVEELARLYDSMEVIGVTCLGSTHAILSRTTFDMCIVDEATQVLQCTVLRPLFAARRFLLVGDPKQLPPIVKSRAARRLGMEESLFHRLMDEEATCTLHLQYRMNQAITDVANQVAYDSRLKCANGQVALATLSIDMQKLSSKTEWLKRACSPEPEDAVLFLDMKSTNVFDFENESRSCRNKAEACLVLDLMECFKEGGVSSKNIGIIAPYREQVSLLRRALASYSVEVNTVDQFQGRDKSILIYSCTKWNVSEDHKVKEGEILNDDRRLAVSVTRAKHKFIIVGNSKALHCYVPLQRLIRASATVSLNDDIVKCLYHKYKEIMI
Summary
Description
O-methyltransferase that catalyzes the 2 O-methylation steps in the ubiquinone biosynthetic pathway.
Key enzyme involved in DNA replication and DNA repair in nucleus and mitochondrion. Involved in Okazaki fragments processing by cleaving long flaps that escape FEN1: flaps that are longer than 27 nucleotides are coated by replication protein A complex (RPA), leading to recruit DNA2 which cleaves the flap until it is too short to bind RPA and becomes a substrate for FEN1. Also involved in 5'-end resection of DNA during double-strand break (DSB) repair: recruited by BLM and mediates the cleavage of 5'-ssDNA, while the 3'-ssDNA cleavage is prevented by the presence of RPA. Also involved in DNA replication checkpoint independently of Okazaki fragments processing. Possesses different enzymatic activities, such as single-stranded DNA (ssDNA)-dependent ATPase, 5'-3' helicase and endonuclease activities. While the ATPase and endonuclease activities are well-defined and play a key role in Okazaki fragments processing and DSB repair, the 5'-3' DNA helicase activity is subject to debate. According to various reports, the helicase activity is weak and its function remains largely unclear. Helicase activity may promote the motion of DNA2 on the flap, helping the nuclease function (By similarity).
Key enzyme involved in DNA replication and DNA repair in nucleus and mitochondrion. Involved in Okazaki fragments processing by cleaving long flaps that escape FEN1: flaps that are longer than 27 nucleotides are coated by replication protein A complex (RPA), leading to recruit DNA2 which cleaves the flap until it is too short to bind RPA and becomes a substrate for FEN1. Also involved in 5'-end resection of DNA during double-strand break (DSB) repair: recruited by BLM and mediates the cleavage of 5'-ssDNA, while the 3'-ssDNA cleavage is prevented by the presence of RPA. Also involved in DNA replication checkpoint independently of Okazaki fragments processing. Possesses different enzymatic activities, such as single-stranded DNA (ssDNA)-dependent ATPase, 5'-3' helicase and endonuclease activities. While the ATPase and endonuclease activities are well-defined and play a key role in Okazaki fragments processing and DSB repair, the 5'-3' DNA helicase activity is subject to debate. According to various reports, the helicase activity is weak and its function remains largely unclear. Helicase activity may promote the motion of DNA2 on the flap, helping the nuclease function (By similarity).
Catalytic Activity
3,4-dihydroxy-5-all-trans-polyprenylbenzoate + S-adenosyl-L-methionine = 3-methoxy,4-hydroxy-5-all-trans-polyprenylbenzoate + H(+) + S-adenosyl-L-homocysteine
a 3-demethylubiquinol + S-adenosyl-L-methionine = a ubiquinol + H(+) + S-adenosyl-L-homocysteine
ATP + H2O = ADP + H(+) + phosphate
a 3-demethylubiquinol + S-adenosyl-L-methionine = a ubiquinol + H(+) + S-adenosyl-L-homocysteine
ATP + H2O = ADP + H(+) + phosphate
Cofactor
[4Fe-4S] cluster
Subunit
Component of a multi-subunit COQ enzyme complex.
Interacts with BLM and WDHD1.
Interacts with BLM and WDHD1.
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. UbiG/COQ3 family.
Belongs to the ubiquitin-conjugating enzyme family.
Belongs to the DNA2/NAM7 helicase family.
Belongs to the mitochondrial carrier (TC 2.A.29) family.
Belongs to the ubiquitin-conjugating enzyme family.
Belongs to the DNA2/NAM7 helicase family.
Belongs to the mitochondrial carrier (TC 2.A.29) family.
Keywords
4Fe-4S
Acetylation
ATP-binding
Complete proteome
DNA damage
DNA repair
DNA replication
DNA-binding
Endonuclease
Helicase
Hydrolase
Iron
Iron-sulfur
Metal-binding
Mitochondrion
Multifunctional enzyme
Nuclease
Nucleotide-binding
Nucleus
Reference proteome
Feature
chain DNA replication ATP-dependent helicase/nuclease DNA2
Uniprot
A0A2A4JCZ8
A0A194Q0H0
A0A3S2LN54
A0A212FMP9
A0A2A4JDD4
A0A0N1IHE3
+ More
A0A2J7PC53 A0A1Y1NCJ4 A0A151X2B9 E9IDW7 A0A195EHV9 K7ITF7 A0A1I8MZ52 A0A0J7KRV7 A0A158NK54 A0A195BB26 A0A067QVG1 E2A0A1 A0A0L0C189 D6WDE8 B0W0R8 A0A1Q3EX06 F4W8F0 A0A154PJA2 A0A3L8DF95 A0A026WRT5 B4KFX0 A0A1I8PCE9 A0A0M4EUJ7 A0A0N0U4B4 A0A232F8M5 A0A182H3D2 B4JBN9 A0A182H3D0 B4PXE6 J9K2Q0 A0A0A1X334 A0A1B0FBW8 A0A1B0BJL6 A0A2H8TS85 A0A087ZYC4 A0A1B0A0B5 A0A1S4EV68 Q8IRL9 A0A1A9YQA3 B3NT26 A0A0K8V5L9 Q9W2Z4 A0A2A3E688 A0A1A9ULP3 E8NH96 B4NE19 Q17Q00 Q86D36 A0A1A9W184 B4IDL1 B4LQU0 B4R772 A0A195FBN3 W5JWN0 B3M6K7 W8C4L7 A0A1W4VUZ8 A0A182FLX6 A0A3B0JWE6 A0A1S3GZW9 A0A1J1IZ91 A0A2S2R1P2 Q29FW1 A0A182QR79 E2BTZ6 A0A2S2NQG5 A0A084WF25 Q7QL33 A0A0A9Y4P5 A0A182J4Q1 A0A0P6BUF1 N6U2A8 A0A0P5XRJ9 A0A1W4XWC4 A0A0P6EG31 A0A210PEQ2 A0A1B0CHA0 A0A3B0JRG1 A0A182W7Z1 A0A2J7PC50 A0A0C9Q039 A0A182JR23 A0A182RS86 T1IXE8 A0A0K2U6U7 A0A182YDW8 D3ZG52 A0A182M9V9 A0A293MGI1 A0A3Q0CRQ2 A0A0H2UH92 A0A1B6E2H8
A0A2J7PC53 A0A1Y1NCJ4 A0A151X2B9 E9IDW7 A0A195EHV9 K7ITF7 A0A1I8MZ52 A0A0J7KRV7 A0A158NK54 A0A195BB26 A0A067QVG1 E2A0A1 A0A0L0C189 D6WDE8 B0W0R8 A0A1Q3EX06 F4W8F0 A0A154PJA2 A0A3L8DF95 A0A026WRT5 B4KFX0 A0A1I8PCE9 A0A0M4EUJ7 A0A0N0U4B4 A0A232F8M5 A0A182H3D2 B4JBN9 A0A182H3D0 B4PXE6 J9K2Q0 A0A0A1X334 A0A1B0FBW8 A0A1B0BJL6 A0A2H8TS85 A0A087ZYC4 A0A1B0A0B5 A0A1S4EV68 Q8IRL9 A0A1A9YQA3 B3NT26 A0A0K8V5L9 Q9W2Z4 A0A2A3E688 A0A1A9ULP3 E8NH96 B4NE19 Q17Q00 Q86D36 A0A1A9W184 B4IDL1 B4LQU0 B4R772 A0A195FBN3 W5JWN0 B3M6K7 W8C4L7 A0A1W4VUZ8 A0A182FLX6 A0A3B0JWE6 A0A1S3GZW9 A0A1J1IZ91 A0A2S2R1P2 Q29FW1 A0A182QR79 E2BTZ6 A0A2S2NQG5 A0A084WF25 Q7QL33 A0A0A9Y4P5 A0A182J4Q1 A0A0P6BUF1 N6U2A8 A0A0P5XRJ9 A0A1W4XWC4 A0A0P6EG31 A0A210PEQ2 A0A1B0CHA0 A0A3B0JRG1 A0A182W7Z1 A0A2J7PC50 A0A0C9Q039 A0A182JR23 A0A182RS86 T1IXE8 A0A0K2U6U7 A0A182YDW8 D3ZG52 A0A182M9V9 A0A293MGI1 A0A3Q0CRQ2 A0A0H2UH92 A0A1B6E2H8
Pubmed
26354079
22118469
28004739
21282665
20075255
25315136
+ More
21347285 24845553 20798317 26108605 18362917 19820115 21719571 30249741 24508170 17994087 28648823 26483478 17550304 25830018 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 24495485 15632085 24438588 12364791 14747013 17210077 25401762 23537049 28812685 25244985 15057822 26319212
21347285 24845553 20798317 26108605 18362917 19820115 21719571 30249741 24508170 17994087 28648823 26483478 17550304 25830018 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 24495485 15632085 24438588 12364791 14747013 17210077 25401762 23537049 28812685 25244985 15057822 26319212
EMBL
NWSH01002014
PCG69424.1
KQ459582
KPI98798.1
RSAL01000051
RVE50295.1
+ More
AGBW02007651 OWR54970.1 PCG69423.1 KQ460208 KPJ16616.1 NEVH01027067 PNF13912.1 GEZM01007071 GEZM01007070 GEZM01007069 GEZM01007068 JAV95429.1 KQ982579 KYQ54573.1 GL762535 EFZ21261.1 KQ978881 KYN27838.1 AAZX01006541 LBMM01003817 KMQ93097.1 ADTU01018700 ADTU01018701 KQ976537 KYM81394.1 KK853205 KDR09858.1 GL435563 EFN73136.1 JRES01001015 KNC26103.1 KQ971321 EEZ99558.2 DS231818 EDS41370.1 GFDL01015201 JAV19844.1 GL887908 EGI69441.1 KQ434912 KZC11378.1 QOIP01000009 RLU18862.1 KK107119 EZA58765.1 CH933807 EDW12096.1 CP012528 ALC48772.1 KQ435826 KOX72024.1 NNAY01000651 OXU27176.1 JXUM01107199 KQ565122 KXJ71310.1 CH916368 EDW02974.1 JXUM01107195 KXJ71308.1 CM000162 EDX02897.1 ABLF02025366 GBXI01009199 JAD05093.1 CCAG010009805 JXJN01015446 GFXV01005271 MBW17076.1 AE014298 BT088860 AAN09623.1 ACS78060.1 CH954180 EDV46206.1 GDHF01018429 JAI33885.1 AAF46543.2 KZ288369 PBC26792.1 BT125972 ADX35951.1 CH964239 EDW81988.1 CH477188 EAT48794.1 BT006332 AAP20828.1 CH480830 EDW45669.1 CH940649 EDW63474.1 CM000366 EDX17541.1 KQ981693 KYN37622.1 ADMH02000009 ETN68108.1 CH902618 EDV38657.1 GAMC01005425 GAMC01005424 JAC01132.1 OUUW01000003 SPP78039.1 CVRI01000064 CRL05010.1 GGMS01014069 MBY83272.1 CH379065 EAL31468.1 AXCN02002217 GL450531 EFN80895.1 GGMR01006811 MBY19430.1 ATLV01023287 KE525342 KFB48819.1 AAAB01008026 EAA03204.4 GBHO01015567 JAG28037.1 GDIP01009317 JAM94398.1 APGK01045214 APGK01045215 APGK01045216 KB741032 ENN74756.1 GDIP01069629 JAM34086.1 GDIQ01075733 JAN19004.1 NEDP02076746 OWF34970.1 AJWK01012146 AJWK01012147 SPP78040.1 PNF13913.1 GBYB01007203 JAG76970.1 JH431646 HACA01016075 CDW33436.1 AXCM01003961 GFWV01013501 MAA38230.1 AABR07044935 GEDC01005166 JAS32132.1
AGBW02007651 OWR54970.1 PCG69423.1 KQ460208 KPJ16616.1 NEVH01027067 PNF13912.1 GEZM01007071 GEZM01007070 GEZM01007069 GEZM01007068 JAV95429.1 KQ982579 KYQ54573.1 GL762535 EFZ21261.1 KQ978881 KYN27838.1 AAZX01006541 LBMM01003817 KMQ93097.1 ADTU01018700 ADTU01018701 KQ976537 KYM81394.1 KK853205 KDR09858.1 GL435563 EFN73136.1 JRES01001015 KNC26103.1 KQ971321 EEZ99558.2 DS231818 EDS41370.1 GFDL01015201 JAV19844.1 GL887908 EGI69441.1 KQ434912 KZC11378.1 QOIP01000009 RLU18862.1 KK107119 EZA58765.1 CH933807 EDW12096.1 CP012528 ALC48772.1 KQ435826 KOX72024.1 NNAY01000651 OXU27176.1 JXUM01107199 KQ565122 KXJ71310.1 CH916368 EDW02974.1 JXUM01107195 KXJ71308.1 CM000162 EDX02897.1 ABLF02025366 GBXI01009199 JAD05093.1 CCAG010009805 JXJN01015446 GFXV01005271 MBW17076.1 AE014298 BT088860 AAN09623.1 ACS78060.1 CH954180 EDV46206.1 GDHF01018429 JAI33885.1 AAF46543.2 KZ288369 PBC26792.1 BT125972 ADX35951.1 CH964239 EDW81988.1 CH477188 EAT48794.1 BT006332 AAP20828.1 CH480830 EDW45669.1 CH940649 EDW63474.1 CM000366 EDX17541.1 KQ981693 KYN37622.1 ADMH02000009 ETN68108.1 CH902618 EDV38657.1 GAMC01005425 GAMC01005424 JAC01132.1 OUUW01000003 SPP78039.1 CVRI01000064 CRL05010.1 GGMS01014069 MBY83272.1 CH379065 EAL31468.1 AXCN02002217 GL450531 EFN80895.1 GGMR01006811 MBY19430.1 ATLV01023287 KE525342 KFB48819.1 AAAB01008026 EAA03204.4 GBHO01015567 JAG28037.1 GDIP01009317 JAM94398.1 APGK01045214 APGK01045215 APGK01045216 KB741032 ENN74756.1 GDIP01069629 JAM34086.1 GDIQ01075733 JAN19004.1 NEDP02076746 OWF34970.1 AJWK01012146 AJWK01012147 SPP78040.1 PNF13913.1 GBYB01007203 JAG76970.1 JH431646 HACA01016075 CDW33436.1 AXCM01003961 GFWV01013501 MAA38230.1 AABR07044935 GEDC01005166 JAS32132.1
Proteomes
UP000218220
UP000053268
UP000283053
UP000007151
UP000053240
UP000235965
+ More
UP000075809 UP000078492 UP000002358 UP000095301 UP000036403 UP000005205 UP000078540 UP000027135 UP000000311 UP000037069 UP000007266 UP000002320 UP000007755 UP000076502 UP000279307 UP000053097 UP000009192 UP000095300 UP000092553 UP000053105 UP000215335 UP000069940 UP000249989 UP000001070 UP000002282 UP000007819 UP000092444 UP000092460 UP000005203 UP000092445 UP000000803 UP000092443 UP000008711 UP000242457 UP000078200 UP000007798 UP000008820 UP000091820 UP000001292 UP000008792 UP000000304 UP000078541 UP000000673 UP000007801 UP000192221 UP000069272 UP000268350 UP000085678 UP000183832 UP000001819 UP000075886 UP000008237 UP000030765 UP000007062 UP000075880 UP000019118 UP000192223 UP000242188 UP000092461 UP000075920 UP000075881 UP000075900 UP000076408 UP000002494 UP000075883 UP000189706
UP000075809 UP000078492 UP000002358 UP000095301 UP000036403 UP000005205 UP000078540 UP000027135 UP000000311 UP000037069 UP000007266 UP000002320 UP000007755 UP000076502 UP000279307 UP000053097 UP000009192 UP000095300 UP000092553 UP000053105 UP000215335 UP000069940 UP000249989 UP000001070 UP000002282 UP000007819 UP000092444 UP000092460 UP000005203 UP000092445 UP000000803 UP000092443 UP000008711 UP000242457 UP000078200 UP000007798 UP000008820 UP000091820 UP000001292 UP000008792 UP000000304 UP000078541 UP000000673 UP000007801 UP000192221 UP000069272 UP000268350 UP000085678 UP000183832 UP000001819 UP000075886 UP000008237 UP000030765 UP000007062 UP000075880 UP000019118 UP000192223 UP000242188 UP000092461 UP000075920 UP000075881 UP000075900 UP000076408 UP000002494 UP000075883 UP000189706
Pfam
Interpro
IPR041679
DNA2/NAM7-like_AAA
+ More
IPR027417 P-loop_NTPase
IPR041677 DNA2/NAM7_AAA_11
IPR014808 DNA_replication_fac_Dna2_N
IPR026851 Dna2
IPR011604 Exonuc_phg/RecB_C
IPR022765 Dna2/Cas4_DUF83
IPR014001 Helicase_ATP-bd
IPR037151 AlkB-like_sf
IPR003593 AAA+_ATPase
IPR010233 UbiG_MeTrfase
IPR029063 SAM-dependent_MTases
IPR007248 Mpv17_PMP22
IPR038657 L19_sf
IPR001857 Ribosomal_L19
IPR008991 Translation_prot_SH3-like_sf
IPR023313 UBQ-conjugating_AS
IPR000608 UBQ-conjugat_E2
IPR016135 UBQ-conjugating_enzyme/RWD
IPR002067 Mit_carrier
IPR023395 Mt_carrier_dom_sf
IPR018108 Mitochondrial_sb/sol_carrier
IPR002167 Graves_DC
IPR027417 P-loop_NTPase
IPR041677 DNA2/NAM7_AAA_11
IPR014808 DNA_replication_fac_Dna2_N
IPR026851 Dna2
IPR011604 Exonuc_phg/RecB_C
IPR022765 Dna2/Cas4_DUF83
IPR014001 Helicase_ATP-bd
IPR037151 AlkB-like_sf
IPR003593 AAA+_ATPase
IPR010233 UbiG_MeTrfase
IPR029063 SAM-dependent_MTases
IPR007248 Mpv17_PMP22
IPR038657 L19_sf
IPR001857 Ribosomal_L19
IPR008991 Translation_prot_SH3-like_sf
IPR023313 UBQ-conjugating_AS
IPR000608 UBQ-conjugat_E2
IPR016135 UBQ-conjugating_enzyme/RWD
IPR002067 Mit_carrier
IPR023395 Mt_carrier_dom_sf
IPR018108 Mitochondrial_sb/sol_carrier
IPR002167 Graves_DC
SUPFAM
CDD
ProteinModelPortal
A0A2A4JCZ8
A0A194Q0H0
A0A3S2LN54
A0A212FMP9
A0A2A4JDD4
A0A0N1IHE3
+ More
A0A2J7PC53 A0A1Y1NCJ4 A0A151X2B9 E9IDW7 A0A195EHV9 K7ITF7 A0A1I8MZ52 A0A0J7KRV7 A0A158NK54 A0A195BB26 A0A067QVG1 E2A0A1 A0A0L0C189 D6WDE8 B0W0R8 A0A1Q3EX06 F4W8F0 A0A154PJA2 A0A3L8DF95 A0A026WRT5 B4KFX0 A0A1I8PCE9 A0A0M4EUJ7 A0A0N0U4B4 A0A232F8M5 A0A182H3D2 B4JBN9 A0A182H3D0 B4PXE6 J9K2Q0 A0A0A1X334 A0A1B0FBW8 A0A1B0BJL6 A0A2H8TS85 A0A087ZYC4 A0A1B0A0B5 A0A1S4EV68 Q8IRL9 A0A1A9YQA3 B3NT26 A0A0K8V5L9 Q9W2Z4 A0A2A3E688 A0A1A9ULP3 E8NH96 B4NE19 Q17Q00 Q86D36 A0A1A9W184 B4IDL1 B4LQU0 B4R772 A0A195FBN3 W5JWN0 B3M6K7 W8C4L7 A0A1W4VUZ8 A0A182FLX6 A0A3B0JWE6 A0A1S3GZW9 A0A1J1IZ91 A0A2S2R1P2 Q29FW1 A0A182QR79 E2BTZ6 A0A2S2NQG5 A0A084WF25 Q7QL33 A0A0A9Y4P5 A0A182J4Q1 A0A0P6BUF1 N6U2A8 A0A0P5XRJ9 A0A1W4XWC4 A0A0P6EG31 A0A210PEQ2 A0A1B0CHA0 A0A3B0JRG1 A0A182W7Z1 A0A2J7PC50 A0A0C9Q039 A0A182JR23 A0A182RS86 T1IXE8 A0A0K2U6U7 A0A182YDW8 D3ZG52 A0A182M9V9 A0A293MGI1 A0A3Q0CRQ2 A0A0H2UH92 A0A1B6E2H8
A0A2J7PC53 A0A1Y1NCJ4 A0A151X2B9 E9IDW7 A0A195EHV9 K7ITF7 A0A1I8MZ52 A0A0J7KRV7 A0A158NK54 A0A195BB26 A0A067QVG1 E2A0A1 A0A0L0C189 D6WDE8 B0W0R8 A0A1Q3EX06 F4W8F0 A0A154PJA2 A0A3L8DF95 A0A026WRT5 B4KFX0 A0A1I8PCE9 A0A0M4EUJ7 A0A0N0U4B4 A0A232F8M5 A0A182H3D2 B4JBN9 A0A182H3D0 B4PXE6 J9K2Q0 A0A0A1X334 A0A1B0FBW8 A0A1B0BJL6 A0A2H8TS85 A0A087ZYC4 A0A1B0A0B5 A0A1S4EV68 Q8IRL9 A0A1A9YQA3 B3NT26 A0A0K8V5L9 Q9W2Z4 A0A2A3E688 A0A1A9ULP3 E8NH96 B4NE19 Q17Q00 Q86D36 A0A1A9W184 B4IDL1 B4LQU0 B4R772 A0A195FBN3 W5JWN0 B3M6K7 W8C4L7 A0A1W4VUZ8 A0A182FLX6 A0A3B0JWE6 A0A1S3GZW9 A0A1J1IZ91 A0A2S2R1P2 Q29FW1 A0A182QR79 E2BTZ6 A0A2S2NQG5 A0A084WF25 Q7QL33 A0A0A9Y4P5 A0A182J4Q1 A0A0P6BUF1 N6U2A8 A0A0P5XRJ9 A0A1W4XWC4 A0A0P6EG31 A0A210PEQ2 A0A1B0CHA0 A0A3B0JRG1 A0A182W7Z1 A0A2J7PC50 A0A0C9Q039 A0A182JR23 A0A182RS86 T1IXE8 A0A0K2U6U7 A0A182YDW8 D3ZG52 A0A182M9V9 A0A293MGI1 A0A3Q0CRQ2 A0A0H2UH92 A0A1B6E2H8
PDB
5EAN
E-value=1.24353e-133,
Score=1225
Ontologies
GO
GO:0017108
GO:0043142
GO:0033567
GO:0004386
GO:0005634
GO:0000784
GO:0005737
GO:0003723
GO:0071932
GO:0004395
GO:1990886
GO:0006744
GO:0008425
GO:0008689
GO:0031314
GO:0016021
GO:0005840
GO:0006412
GO:0003735
GO:0005524
GO:0016740
GO:0004518
GO:0000729
GO:0051539
GO:0046872
GO:0000076
GO:0005739
GO:0003677
GO:0003678
GO:0006260
GO:0043139
GO:0005760
GO:0005743
GO:0022857
GO:0043137
GO:0043504
GO:0016890
GO:0042645
GO:0006284
GO:0045740
GO:0006264
PANTHER
Topology
Subcellular location
Mitochondrion inner membrane
Nucleus
Mitochondrion
Nucleus
Mitochondrion
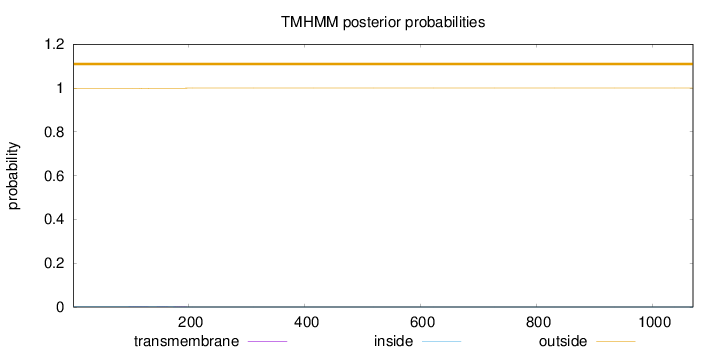
Length:
1070
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0366300000000002
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00184
outside
1 - 1070
Population Genetic Test Statistics
Pi
287.183875
Theta
190.066637
Tajima's D
1.843364
CLR
0.134595
CSRT
0.855807209639518
Interpretation
Uncertain
