Pre Gene Modal
BGIBMGA012982
Annotation
putative_methyltransferase_WBSCR22-like_protein_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.391 Mitochondrial Reliability : 1.581
Sequence
CDS
ATGACTGAGAGATGCTTAGAATTACTTCTATTACCGGAAGATTCATCTTGCTTGTTGTTGGATATTGGATGTGGTTCAGGATTATCTGGTACAGTGCTAGAAGAAAATGGACATATGTGGATTGGGATGGACATATCATCTTCAATGCTTGATGTTGCTGTAGAGAGAGACACTGAAGGTGGTTTAGTTCTGGCTGATATGGGTGAAGGTGTCCCGTTTAGAGCAGGTTGCTTTGATGGAGCAGTATCCGTATCAGCTATACAATGGTTATTTAATGCTGATAAGAAAACTCATAATCCAGTGAAAAGACTAAACAAATTCTTCACCACTTTATATTCAAGTTTGTCAAGGTCTGCTCGAGCTGTCTTCCAATTTTACCCTGAGAATGAGAAGCAATTAGAACTCCTAACGACACAGGCAATGAAAGCTGGCTTTTATGGAGGAGTTGTAATTGACTATCCAAATTCTGCCAAAGCAAAGAAGTTCTTTTTAGTTTTAATGACTGGTGGCGTTGGTCCTTTACCTCAGGCTTTAGGTACTGAAGAACCAGACAGTCTTCAAGTTAAATATGCTAAAAGAGAAGCTACAAAAGGGATAAGAGGAAAACCTATAAAGAACTCAAAAGACTGGATTTTGGAGAAGAAAGAACGAAGAAGGAAACAGGGCAAAATTACAAAACCTGACACTAAATACACAGGAAGAAAAAGAAGTGGTCGATTCTAA
Protein
MTERCLELLLLPEDSSCLLLDIGCGSGLSGTVLEENGHMWIGMDISSSMLDVAVERDTEGGLVLADMGEGVPFRAGCFDGAVSVSAIQWLFNADKKTHNPVKRLNKFFTTLYSSLSRSARAVFQFYPENEKQLELLTTQAMKAGFYGGVVIDYPNSAKAKKFFLVLMTGGVGPLPQALGTEEPDSLQVKYAKREATKGIRGKPIKNSKDWILEKKERRRKQGKITKPDTKYTGRKRSGRF
Summary
Uniprot
G8GE16
H9JTX0
A0A2A4JDC9
A0A2H1W1V4
S4P8W2
A0A194PZP1
+ More
A0A0N1IIM7 A0A0L7LT43 A0A212FMK7 A0A437BIM3 A0A1I8M8X2 T1PM71 D2A4C3 A0A1I8NQV5 A0A1B0CL63 A0A182LAZ1 A0T4G1 A0A182I909 A0A1L8DAT0 A0A182WVC8 A0A182UY51 A0A182TP61 A0A182H7I2 A0A0A9ZB46 A0A336KJ26 A0A026WUC2 A0A182K4L4 A0A146KNG6 Q16RH3 A0A1Y1LBP7 A0A182PKG3 A0A154PN83 A0A2M4AM51 A0A2M4AMZ8 W5JCQ9 A0A1Q3F4M8 A0A2M4BWL0 A0A2M4BW67 A0A1B3PDA1 A0A182F941 A0A2M3ZBJ3 A0A2M4BWE4 A0A1Q3F4S9 A0A1B6LPZ3 A0A1W4WJX6 A0A1B3PDL9 A0A182J2T7 A0A1B0A382 A0A1J1IU19 A0A1B0DPD6 B0W3K8 A0A1A9VFL5 A0A151XDN8 D3TN55 Q295H9 A0A3B0KCZ2 B3LXB5 B4GDV1 A0A1B6GS34 A0A2R7WWP7 A0A1B6HP56 Q9VHW6 B4PQX4 E1ZX81 F4WAR3 E2C601 A0A182QR13 A0A151IES8 A0A182N4K8 B4R0D5 B4I3D8 A0A1W4VL60 J3JTD7 B4NJ38 A0A195F130 U4U0E7 B4JUS1 B3NYV1 B4M571 W8BUW9 B4KDE8 A0A182SNI8 A0A0K8V7M7 A0A158P3J1 A0A182VY18 A0A151JQX1 A0A182LVY7 A0A182R5Q2 A0A0V0GCZ4 A0A034WVC1 A0A2J7RBF9 A0A182YP32 A0A2R4SE39 A0A195BWW0 R4G8D8 A0A084W4Z0 A0A310SBN5 A0A0M4F6A7 A0A0J7NLF8 A0A0P4VSW9
A0A0N1IIM7 A0A0L7LT43 A0A212FMK7 A0A437BIM3 A0A1I8M8X2 T1PM71 D2A4C3 A0A1I8NQV5 A0A1B0CL63 A0A182LAZ1 A0T4G1 A0A182I909 A0A1L8DAT0 A0A182WVC8 A0A182UY51 A0A182TP61 A0A182H7I2 A0A0A9ZB46 A0A336KJ26 A0A026WUC2 A0A182K4L4 A0A146KNG6 Q16RH3 A0A1Y1LBP7 A0A182PKG3 A0A154PN83 A0A2M4AM51 A0A2M4AMZ8 W5JCQ9 A0A1Q3F4M8 A0A2M4BWL0 A0A2M4BW67 A0A1B3PDA1 A0A182F941 A0A2M3ZBJ3 A0A2M4BWE4 A0A1Q3F4S9 A0A1B6LPZ3 A0A1W4WJX6 A0A1B3PDL9 A0A182J2T7 A0A1B0A382 A0A1J1IU19 A0A1B0DPD6 B0W3K8 A0A1A9VFL5 A0A151XDN8 D3TN55 Q295H9 A0A3B0KCZ2 B3LXB5 B4GDV1 A0A1B6GS34 A0A2R7WWP7 A0A1B6HP56 Q9VHW6 B4PQX4 E1ZX81 F4WAR3 E2C601 A0A182QR13 A0A151IES8 A0A182N4K8 B4R0D5 B4I3D8 A0A1W4VL60 J3JTD7 B4NJ38 A0A195F130 U4U0E7 B4JUS1 B3NYV1 B4M571 W8BUW9 B4KDE8 A0A182SNI8 A0A0K8V7M7 A0A158P3J1 A0A182VY18 A0A151JQX1 A0A182LVY7 A0A182R5Q2 A0A0V0GCZ4 A0A034WVC1 A0A2J7RBF9 A0A182YP32 A0A2R4SE39 A0A195BWW0 R4G8D8 A0A084W4Z0 A0A310SBN5 A0A0M4F6A7 A0A0J7NLF8 A0A0P4VSW9
Pubmed
19121390
23622113
26354079
26227816
22118469
25315136
+ More
18362917 19820115 20966253 12364791 14747013 17210077 26483478 25401762 24508170 30249741 26823975 17510324 28004739 20920257 23761445 20353571 15632085 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20798317 21719571 22516182 23537049 18057021 24495485 21347285 25348373 25244985 24438588 27129103
18362917 19820115 20966253 12364791 14747013 17210077 26483478 25401762 24508170 30249741 26823975 17510324 28004739 20920257 23761445 20353571 15632085 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20798317 21719571 22516182 23537049 18057021 24495485 21347285 25348373 25244985 24438588 27129103
EMBL
JN127349
AET05795.1
BABH01003711
NWSH01002014
PCG69422.1
ODYU01005809
+ More
SOQ47070.1 GAIX01004009 JAA88551.1 KQ459582 KPI98797.1 KQ460208 KPJ16615.1 JTDY01000139 KOB78633.1 AGBW02007651 OWR54971.1 RSAL01000051 RVE50294.1 KA648993 AFP63622.1 KQ971348 EFA04808.1 AJWK01016952 AJWK01016953 AJWK01016954 AAAB01008844 EAA05981.2 APCN01003147 GFDF01010503 JAV03581.1 JXUM01028280 KQ560816 KXJ80774.1 GBHO01003026 GBRD01001131 JAG40578.1 JAG64690.1 UFQS01000501 UFQT01000501 SSX04447.1 SSX24811.1 KK107109 QOIP01000008 EZA59261.1 RLU19935.1 GDHC01021772 JAP96856.1 CH477709 EAT37016.1 GEZM01061828 JAV70301.1 KQ434998 KZC13341.1 GGFK01008542 MBW41863.1 GGFK01008657 MBW41978.1 ADMH02001893 ETN60614.1 GFDL01012607 JAV22438.1 GGFJ01008180 MBW57321.1 GGFJ01008176 MBW57317.1 KU659909 AOG17708.1 GGFM01005077 MBW25828.1 GGFJ01008181 MBW57322.1 GFDL01012508 JAV22537.1 GEBQ01014224 JAT25753.1 KU660036 AOG17834.1 CVRI01000057 CRL02025.1 AJVK01018247 DS231832 EDS31794.1 KQ982268 KYQ58496.1 CCAG010010183 EZ422857 ADD19133.1 CM000070 EAL28733.1 OUUW01000007 SPP83586.1 CH902617 EDV42759.1 CH479182 EDW34612.1 GECZ01004532 JAS65237.1 KK855815 PTY23969.1 GECU01031248 JAS76458.1 AE014297 AY071172 AAF54183.1 AAL48794.1 CM000160 EDW96298.1 GL435030 EFN74171.1 GL888053 EGI68669.1 GL452895 EFN76606.1 AXCN02000044 KQ977858 KYM99330.1 CM000364 EDX12060.1 CH480821 EDW55302.1 APGK01054968 BT126493 KB741256 AEE61457.1 ENN71752.1 CH964272 EDW83831.1 KRF99897.1 KQ981880 KYN33877.1 KB631749 ERL85773.1 CH916374 EDV91241.1 CH954181 EDV48214.1 CH940652 EDW59782.1 GAMC01009469 GAMC01009468 GAMC01009467 JAB97086.1 CH933806 EDW15957.1 GDHF01017448 JAI34866.1 ADTU01008114 KQ978621 KYN29742.1 AXCM01004998 GECL01000384 JAP05740.1 GAKP01001259 JAC57693.1 NEVH01005904 PNF38169.1 MF101772 AVZ46909.1 KQ976394 KYM93119.1 ACPB03013747 GAHY01001147 JAA76363.1 ATLV01020424 KE525302 KFB45284.1 KQ761783 OAD57060.1 CP012526 ALC47207.1 LBMM01003626 KMQ93310.1 GDKW01001525 JAI55070.1
SOQ47070.1 GAIX01004009 JAA88551.1 KQ459582 KPI98797.1 KQ460208 KPJ16615.1 JTDY01000139 KOB78633.1 AGBW02007651 OWR54971.1 RSAL01000051 RVE50294.1 KA648993 AFP63622.1 KQ971348 EFA04808.1 AJWK01016952 AJWK01016953 AJWK01016954 AAAB01008844 EAA05981.2 APCN01003147 GFDF01010503 JAV03581.1 JXUM01028280 KQ560816 KXJ80774.1 GBHO01003026 GBRD01001131 JAG40578.1 JAG64690.1 UFQS01000501 UFQT01000501 SSX04447.1 SSX24811.1 KK107109 QOIP01000008 EZA59261.1 RLU19935.1 GDHC01021772 JAP96856.1 CH477709 EAT37016.1 GEZM01061828 JAV70301.1 KQ434998 KZC13341.1 GGFK01008542 MBW41863.1 GGFK01008657 MBW41978.1 ADMH02001893 ETN60614.1 GFDL01012607 JAV22438.1 GGFJ01008180 MBW57321.1 GGFJ01008176 MBW57317.1 KU659909 AOG17708.1 GGFM01005077 MBW25828.1 GGFJ01008181 MBW57322.1 GFDL01012508 JAV22537.1 GEBQ01014224 JAT25753.1 KU660036 AOG17834.1 CVRI01000057 CRL02025.1 AJVK01018247 DS231832 EDS31794.1 KQ982268 KYQ58496.1 CCAG010010183 EZ422857 ADD19133.1 CM000070 EAL28733.1 OUUW01000007 SPP83586.1 CH902617 EDV42759.1 CH479182 EDW34612.1 GECZ01004532 JAS65237.1 KK855815 PTY23969.1 GECU01031248 JAS76458.1 AE014297 AY071172 AAF54183.1 AAL48794.1 CM000160 EDW96298.1 GL435030 EFN74171.1 GL888053 EGI68669.1 GL452895 EFN76606.1 AXCN02000044 KQ977858 KYM99330.1 CM000364 EDX12060.1 CH480821 EDW55302.1 APGK01054968 BT126493 KB741256 AEE61457.1 ENN71752.1 CH964272 EDW83831.1 KRF99897.1 KQ981880 KYN33877.1 KB631749 ERL85773.1 CH916374 EDV91241.1 CH954181 EDV48214.1 CH940652 EDW59782.1 GAMC01009469 GAMC01009468 GAMC01009467 JAB97086.1 CH933806 EDW15957.1 GDHF01017448 JAI34866.1 ADTU01008114 KQ978621 KYN29742.1 AXCM01004998 GECL01000384 JAP05740.1 GAKP01001259 JAC57693.1 NEVH01005904 PNF38169.1 MF101772 AVZ46909.1 KQ976394 KYM93119.1 ACPB03013747 GAHY01001147 JAA76363.1 ATLV01020424 KE525302 KFB45284.1 KQ761783 OAD57060.1 CP012526 ALC47207.1 LBMM01003626 KMQ93310.1 GDKW01001525 JAI55070.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000037510
UP000007151
+ More
UP000283053 UP000095301 UP000007266 UP000095300 UP000092461 UP000075882 UP000007062 UP000075840 UP000076407 UP000075903 UP000075902 UP000069940 UP000249989 UP000053097 UP000279307 UP000075881 UP000008820 UP000075885 UP000076502 UP000000673 UP000069272 UP000192223 UP000075880 UP000092445 UP000183832 UP000092462 UP000002320 UP000078200 UP000075809 UP000092444 UP000001819 UP000268350 UP000007801 UP000008744 UP000000803 UP000002282 UP000000311 UP000007755 UP000008237 UP000075886 UP000078542 UP000075884 UP000000304 UP000001292 UP000192221 UP000019118 UP000007798 UP000078541 UP000030742 UP000001070 UP000008711 UP000008792 UP000009192 UP000075901 UP000005205 UP000075920 UP000078492 UP000075883 UP000075900 UP000235965 UP000076408 UP000078540 UP000015103 UP000030765 UP000092553 UP000036403
UP000283053 UP000095301 UP000007266 UP000095300 UP000092461 UP000075882 UP000007062 UP000075840 UP000076407 UP000075903 UP000075902 UP000069940 UP000249989 UP000053097 UP000279307 UP000075881 UP000008820 UP000075885 UP000076502 UP000000673 UP000069272 UP000192223 UP000075880 UP000092445 UP000183832 UP000092462 UP000002320 UP000078200 UP000075809 UP000092444 UP000001819 UP000268350 UP000007801 UP000008744 UP000000803 UP000002282 UP000000311 UP000007755 UP000008237 UP000075886 UP000078542 UP000075884 UP000000304 UP000001292 UP000192221 UP000019118 UP000007798 UP000078541 UP000030742 UP000001070 UP000008711 UP000008792 UP000009192 UP000075901 UP000005205 UP000075920 UP000078492 UP000075883 UP000075900 UP000235965 UP000076408 UP000078540 UP000015103 UP000030765 UP000092553 UP000036403
Pfam
Interpro
Gene 3D
ProteinModelPortal
G8GE16
H9JTX0
A0A2A4JDC9
A0A2H1W1V4
S4P8W2
A0A194PZP1
+ More
A0A0N1IIM7 A0A0L7LT43 A0A212FMK7 A0A437BIM3 A0A1I8M8X2 T1PM71 D2A4C3 A0A1I8NQV5 A0A1B0CL63 A0A182LAZ1 A0T4G1 A0A182I909 A0A1L8DAT0 A0A182WVC8 A0A182UY51 A0A182TP61 A0A182H7I2 A0A0A9ZB46 A0A336KJ26 A0A026WUC2 A0A182K4L4 A0A146KNG6 Q16RH3 A0A1Y1LBP7 A0A182PKG3 A0A154PN83 A0A2M4AM51 A0A2M4AMZ8 W5JCQ9 A0A1Q3F4M8 A0A2M4BWL0 A0A2M4BW67 A0A1B3PDA1 A0A182F941 A0A2M3ZBJ3 A0A2M4BWE4 A0A1Q3F4S9 A0A1B6LPZ3 A0A1W4WJX6 A0A1B3PDL9 A0A182J2T7 A0A1B0A382 A0A1J1IU19 A0A1B0DPD6 B0W3K8 A0A1A9VFL5 A0A151XDN8 D3TN55 Q295H9 A0A3B0KCZ2 B3LXB5 B4GDV1 A0A1B6GS34 A0A2R7WWP7 A0A1B6HP56 Q9VHW6 B4PQX4 E1ZX81 F4WAR3 E2C601 A0A182QR13 A0A151IES8 A0A182N4K8 B4R0D5 B4I3D8 A0A1W4VL60 J3JTD7 B4NJ38 A0A195F130 U4U0E7 B4JUS1 B3NYV1 B4M571 W8BUW9 B4KDE8 A0A182SNI8 A0A0K8V7M7 A0A158P3J1 A0A182VY18 A0A151JQX1 A0A182LVY7 A0A182R5Q2 A0A0V0GCZ4 A0A034WVC1 A0A2J7RBF9 A0A182YP32 A0A2R4SE39 A0A195BWW0 R4G8D8 A0A084W4Z0 A0A310SBN5 A0A0M4F6A7 A0A0J7NLF8 A0A0P4VSW9
A0A0N1IIM7 A0A0L7LT43 A0A212FMK7 A0A437BIM3 A0A1I8M8X2 T1PM71 D2A4C3 A0A1I8NQV5 A0A1B0CL63 A0A182LAZ1 A0T4G1 A0A182I909 A0A1L8DAT0 A0A182WVC8 A0A182UY51 A0A182TP61 A0A182H7I2 A0A0A9ZB46 A0A336KJ26 A0A026WUC2 A0A182K4L4 A0A146KNG6 Q16RH3 A0A1Y1LBP7 A0A182PKG3 A0A154PN83 A0A2M4AM51 A0A2M4AMZ8 W5JCQ9 A0A1Q3F4M8 A0A2M4BWL0 A0A2M4BW67 A0A1B3PDA1 A0A182F941 A0A2M3ZBJ3 A0A2M4BWE4 A0A1Q3F4S9 A0A1B6LPZ3 A0A1W4WJX6 A0A1B3PDL9 A0A182J2T7 A0A1B0A382 A0A1J1IU19 A0A1B0DPD6 B0W3K8 A0A1A9VFL5 A0A151XDN8 D3TN55 Q295H9 A0A3B0KCZ2 B3LXB5 B4GDV1 A0A1B6GS34 A0A2R7WWP7 A0A1B6HP56 Q9VHW6 B4PQX4 E1ZX81 F4WAR3 E2C601 A0A182QR13 A0A151IES8 A0A182N4K8 B4R0D5 B4I3D8 A0A1W4VL60 J3JTD7 B4NJ38 A0A195F130 U4U0E7 B4JUS1 B3NYV1 B4M571 W8BUW9 B4KDE8 A0A182SNI8 A0A0K8V7M7 A0A158P3J1 A0A182VY18 A0A151JQX1 A0A182LVY7 A0A182R5Q2 A0A0V0GCZ4 A0A034WVC1 A0A2J7RBF9 A0A182YP32 A0A2R4SE39 A0A195BWW0 R4G8D8 A0A084W4Z0 A0A310SBN5 A0A0M4F6A7 A0A0J7NLF8 A0A0P4VSW9
PDB
6G4W
E-value=9.55164e-68,
Score=649
Ontologies
GO
PANTHER
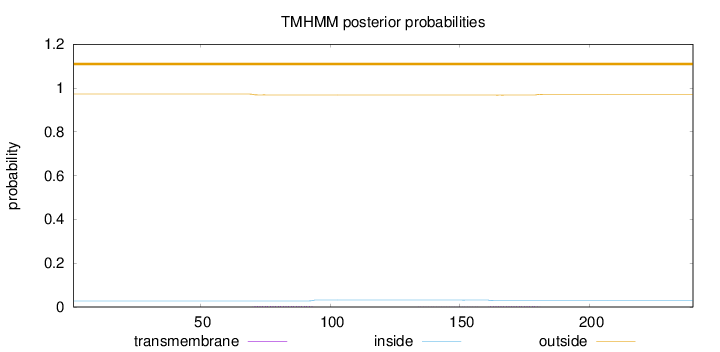
Topology
Length:
240
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.16815
Exp number, first 60 AAs:
0.00165
Total prob of N-in:
0.02734
outside
1 - 240
Population Genetic Test Statistics
Pi
212.781128
Theta
181.200802
Tajima's D
0.62905
CLR
0.056086
CSRT
0.55727213639318
Interpretation
Uncertain
