Gene
KWMTBOMO09625 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012983
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_ATP-dependent_RNA_helicase_Ddx1-like_[Bombyx_mori]
Full name
ATP-dependent RNA helicase Ddx1
Location in the cell
Cytoplasmic Reliability : 3.104
Sequence
CDS
ATGACTGCTTTCGAAGAATTTGGTGTGCTGCCTGAAATAGGCAAAGCTATTGAAGAAATGGATTGGACTCTACCGACTGATGTCCAAGCCGAAGCTATTCCCCTAATCTTAGGCGGTGGAGATGTACTTATGGCAGCCGAAACTGGCAGTGGAAAGACAGGAGCATTCTGTTTACCTATATTGCAGATTGTCTGGGAAACCTTGAAAGATATGCAGGAAGGAAAAACCGGTAAAGTAATAACACAAACGTCTTCTGAGTGTACTATGTCATTTTTCGATCGCACTGACGCACTGGCTGTTACTCCTGACGGGTTACGGTGTCAATCCAGAAATCAAAAAGACTGGCACGGGTGTAGGGCAACCAAAGGTGTTCATACGAAAGGTGCATATTACTTTGAAGCCACTGTCACTGATGAGGGCTTATGCCGTGTCGGCTGGTCTACACAAGCGGCAAAGCTGGATCTTGGAACTGACAGGCTAGGCTATGGATTTGGTGGTACAGGCAAGAAATCAAATTGTAAGCAATTTGATGATTATGGCGAGGCTTATGGGATGAATGATGTCATTGGCTGTTTCCTGAATTTAAATAATGGTGAAATTCGATACTCTAAAAACGGAGAAGACTTGGGTGTGGCTTTCAGTCTTGATTCATCTAGAAAGTCAGACTGTTACTTCCCTGCAGTTGTCTTGAAAAATGCTGAGATGAGTTTTAACTTTGGTGCCACACCATTTAAGTACCCACCACCAAACGGATACATAGCAATCAGTCAAGCACCCAAAGAAAACGTCAAGCAGAATGTTATAGCCGCTGTAGCTTCAACAGATATTAAACCAGTGAATAATGCACCGCAGGCTATTATTATTGAGCCGTCCAGAGAATTAGCGGAGCAGACCTGTACACAATTTACGAAATTCAAGAAGTATTTAGAAAATCCCAAAATTCGGGAGCTGCTGGTTGTTGGAGGTATAAATGTTAAAGATCAAATTAACCAGTTGAACTGTGGGGTGGATGTCGTCGTTGGGACGCCGGGACGCTTGGAAGACCTTATCCAAGGAGGATACTTATCACTGACGCATTGCCGTTTCTTCGTCTTGGACGAGGCTGATGGTCTCTTGAAAGCGGGTTACGGAGAACTGATTGAACGCCTCCATCGTCAGATCCCGAAGATAACATCCGATGGACGGAGACTGCAGATGGTTGTGTGTTCGGCAACCCTGCACGCATTCGAAGTCAAGAAAATGGCTGAAAAATTAATGCACTTTCCGACTTGGGTCGATTTGAAAGGGGAAGACGCAGTTCCTGAGACCGTACACCATGTTGTTGTGAACGTAGACCCGCAAAAGGACAGCTCTTGGCAAACACTGCGCAAGCACGTGCAGACCGACGGTGTGCACGCTAAGGACAACATTCGAAGCGGCAACACTACGCCTGAAACGTTGTCTGAAGCTGTTAAGATACTGAAAGGAGAATACTGTGTGCGCGCCATCAGGGAACACAAAATGGACCGGGCCATAATATTCTGCCGCACTAAATTGGATTGCGACAACCTCGAGCGGTACTTCAAAACGTTCGGCAATGAGCTGTCCTGTGTGTGCCTGCACGGCGACCGAAAACCAAAGGAGCGCAAAGACAATTTGGAGATATTCAAGAACAGCCAAGTGAAGTTTTTGATTTGCACCGACGTCGCTGCTCGCGGCATCGACATCTCGGGCCTGCCGTTCATGATCAACATAACGCTTCCCGACGAAAAGTCCAACTACGTACATCGCATCGGGCGCGTCGGTCGCGCGGAACGCATGGGCCTGGCCATCAGTCTCGTCTCCACCGTACCCGAAAAGGTATGGTATCATGGCGAGTGGTGTTCATCTCGCGGACGGAACTGCTGGAACACTAATCTGATTGACGACAAACCTAAGGGATGCTGCATGTGGTACAATGAACCTCAGTACCTGGCGGATATTGAAGATCACCTAAATATCACGATACAGCAGGTCGAGACGGATATGAAAGTCCCATCCAATGAGTTTGACGGTAAAGTAGTTTACGGTGAAAAAAGAAAACAAGCTGGTTCCGGATACCAAGACCATGTCAGTCAAATGGCGCCTGTTGTGAAATCTCTGCAAGAGCTAGAGTCGCAAGCACAACTCTACTATCTTAAAATGCATCATAATGTCTCCATTTCTTGA
Protein
MTAFEEFGVLPEIGKAIEEMDWTLPTDVQAEAIPLILGGGDVLMAAETGSGKTGAFCLPILQIVWETLKDMQEGKTGKVITQTSSECTMSFFDRTDALAVTPDGLRCQSRNQKDWHGCRATKGVHTKGAYYFEATVTDEGLCRVGWSTQAAKLDLGTDRLGYGFGGTGKKSNCKQFDDYGEAYGMNDVIGCFLNLNNGEIRYSKNGEDLGVAFSLDSSRKSDCYFPAVVLKNAEMSFNFGATPFKYPPPNGYIAISQAPKENVKQNVIAAVASTDIKPVNNAPQAIIIEPSRELAEQTCTQFTKFKKYLENPKIRELLVVGGINVKDQINQLNCGVDVVVGTPGRLEDLIQGGYLSLTHCRFFVLDEADGLLKAGYGELIERLHRQIPKITSDGRRLQMVVCSATLHAFEVKKMAEKLMHFPTWVDLKGEDAVPETVHHVVVNVDPQKDSSWQTLRKHVQTDGVHAKDNIRSGNTTPETLSEAVKILKGEYCVRAIREHKMDRAIIFCRTKLDCDNLERYFKTFGNELSCVCLHGDRKPKERKDNLEIFKNSQVKFLICTDVAARGIDISGLPFMINITLPDEKSNYVHRIGRVGRAERMGLAISLVSTVPEKVWYHGEWCSSRGRNCWNTNLIDDKPKGCCMWYNEPQYLADIEDHLNITIQQVETDMKVPSNEFDGKVVYGEKRKQAGSGYQDHVSQMAPVVKSLQELESQAQLYYLKMHHNVSIS
Summary
Description
Acts as an ATP-dependent RNA helicase, able to unwind both RNA-RNA and RNA-DNA duplexes. Possesses 5' single-stranded RNA overhang nuclease activity (By similarity).
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Similarity
Belongs to the DEAD box helicase family. DDX1 subfamily.
Keywords
ATP-binding
Complete proteome
Exonuclease
Helicase
Hydrolase
Nuclease
Nucleotide-binding
Reference proteome
RNA-binding
Feature
chain ATP-dependent RNA helicase Ddx1
Uniprot
A0A0N1IKA5
A0A437BIM4
A0A2A4K0C1
A0A2H1W1Y1
A0A212FML7
A0A2J7PVU4
+ More
A0A067RH28 A0A087ZYP9 A0A026W4Y4 A0A2A3EEI5 A0A2Z5U645 A0A2J7PVU0 A0A158NYE9 E2BW30 F4WT60 K7J0U0 A0A195F6A1 A0A232EWQ9 A0A195AYX2 A0A151J561 A0A1B6IKZ6 E2ARV8 A0A0C9R6F3 A0A0T6B0Q4 A0A195D0L0 A0A0N1ITN7 A0A0J7KSE3 A0A182R027 D6WQ19 A0A182G367 A0A0L7RE09 E0W1E6 A0A182K800 A0A182HXI7 A0A182X1S6 A0A182LI74 Q7PMT7 A0A1B0GKA8 E9JD76 A0A182RXG1 A0A182UUN4 A0A1Q3FBR9 Q17I24 W8AWI1 A0A1S4F242 A0A182TVK1 A0A1Y1L3E8 A0A310SMN5 A0A1A9WH97 A0A1J1HPU2 S4PXG6 A0A182NNC6 A0A182M988 A0A1W4XKF0 A0A1A9UP81 A0A1B0FBQ6 A0A1A9XK42 A0A1B0ANP0 A0A1A9ZBX0 A0A336MTA5 A0A1I8M934 A0A034VY24 A0A182IR87 A0A1W4XA81 A0A0K8WAU6 A0A182WCL7 N6UFM6 A0A2M4AMX9 W5JRR2 A0A084VZR8 A0A1I8PRU1 A0A1Z5KW41 A0A182FLR0 A0A2R5LJG8 A0A182SHG1 A0A2P8XK11 A0A1W4UZ38 A0A182YI26 B4LE09 B4IXM8 B4PI64 B4IAZ3 A0A0L0CMG3 Q9VNV3 A0A1D2MD94 A0A0J9S022 T1J2C5 B3MAL1 B4KVA3 B4MN98 A0A023GNS0 A0A131YXC4 B3NJ32 A0A224YI64 A0A2P2HX25 B4H8N8 A0A0R3P8Y6 Q29DM1 A0A226DHY8 A0A3B0JSR4
A0A067RH28 A0A087ZYP9 A0A026W4Y4 A0A2A3EEI5 A0A2Z5U645 A0A2J7PVU0 A0A158NYE9 E2BW30 F4WT60 K7J0U0 A0A195F6A1 A0A232EWQ9 A0A195AYX2 A0A151J561 A0A1B6IKZ6 E2ARV8 A0A0C9R6F3 A0A0T6B0Q4 A0A195D0L0 A0A0N1ITN7 A0A0J7KSE3 A0A182R027 D6WQ19 A0A182G367 A0A0L7RE09 E0W1E6 A0A182K800 A0A182HXI7 A0A182X1S6 A0A182LI74 Q7PMT7 A0A1B0GKA8 E9JD76 A0A182RXG1 A0A182UUN4 A0A1Q3FBR9 Q17I24 W8AWI1 A0A1S4F242 A0A182TVK1 A0A1Y1L3E8 A0A310SMN5 A0A1A9WH97 A0A1J1HPU2 S4PXG6 A0A182NNC6 A0A182M988 A0A1W4XKF0 A0A1A9UP81 A0A1B0FBQ6 A0A1A9XK42 A0A1B0ANP0 A0A1A9ZBX0 A0A336MTA5 A0A1I8M934 A0A034VY24 A0A182IR87 A0A1W4XA81 A0A0K8WAU6 A0A182WCL7 N6UFM6 A0A2M4AMX9 W5JRR2 A0A084VZR8 A0A1I8PRU1 A0A1Z5KW41 A0A182FLR0 A0A2R5LJG8 A0A182SHG1 A0A2P8XK11 A0A1W4UZ38 A0A182YI26 B4LE09 B4IXM8 B4PI64 B4IAZ3 A0A0L0CMG3 Q9VNV3 A0A1D2MD94 A0A0J9S022 T1J2C5 B3MAL1 B4KVA3 B4MN98 A0A023GNS0 A0A131YXC4 B3NJ32 A0A224YI64 A0A2P2HX25 B4H8N8 A0A0R3P8Y6 Q29DM1 A0A226DHY8 A0A3B0JSR4
EC Number
3.6.4.13
Pubmed
26354079
22118469
24845553
24508170
26760975
21347285
+ More
20798317 21719571 20075255 28648823 18362917 19820115 26483478 20566863 20966253 12364791 14747013 17210077 21282665 17510324 24495485 28004739 23622113 25315136 25348373 23537049 20920257 23761445 24438588 28528879 29403074 25244985 17994087 17550304 26108605 8666277 10731132 12537572 12537569 27289101 22936249 26830274 28797301 15632085
20798317 21719571 20075255 28648823 18362917 19820115 26483478 20566863 20966253 12364791 14747013 17210077 21282665 17510324 24495485 28004739 23622113 25315136 25348373 23537049 20920257 23761445 24438588 28528879 29403074 25244985 17994087 17550304 26108605 8666277 10731132 12537572 12537569 27289101 22936249 26830274 28797301 15632085
EMBL
LADI01006439
KPJ20646.1
RSAL01000051
RVE50292.1
NWSH01000320
PCG77499.1
+ More
ODYU01005809 SOQ47068.1 AGBW02007651 OWR54973.1 NEVH01020942 PNF20456.1 KK852686 KDR18468.1 KK107419 EZA51117.1 KZ288269 PBC29884.1 FX985851 BBA93738.1 PNF20457.1 ADTU01003779 GL451091 EFN80062.1 GL888331 EGI62673.1 KQ981768 KYN35998.1 NNAY01001834 OXU22765.1 KQ976701 KYM77235.1 KQ980055 KYN17991.1 GECU01020109 GECU01006522 JAS87597.1 JAT01185.1 GL442175 EFN63841.1 GBYB01008382 JAG78149.1 LJIG01016386 KRT80756.1 KQ977004 KYN06453.1 KQ435765 KOX75351.1 LBMM01003779 KMQ93149.1 AXCN02001905 KQ971354 EFA06143.1 JXUM01141022 JXUM01141023 KQ569173 KXJ68738.1 KQ414613 KOC69059.1 DS235870 EEB19452.1 APCN01005308 AAAB01008968 EAA13221.5 AJWK01028622 GL771866 EFZ09293.1 GFDL01010021 JAV25024.1 CH477243 EAT46335.1 GAMC01017427 JAB89128.1 GEZM01067947 JAV67311.1 KQ759870 OAD62402.1 CVRI01000006 CRK88233.1 GAIX01005073 JAA87487.1 AXCM01002532 CCAG010004106 JXJN01000907 UFQS01002152 UFQT01002152 SSX13347.1 SSX32781.1 GAKP01012197 JAC46755.1 GDHF01004021 JAI48293.1 APGK01028591 APGK01028592 APGK01028593 APGK01028594 KB740694 ENN79436.1 GGFK01008822 MBW42143.1 ADMH02000548 ETN65973.1 ATLV01018954 KE525256 KFB43462.1 GFJQ02007725 JAV99244.1 GGLE01005518 MBY09644.1 PYGN01001881 PSN32349.1 CH940647 EDW70052.2 CH916366 EDV97490.1 CM000159 EDW95512.1 CH480826 EDW44456.1 JRES01000296 KNC32659.1 U34773 AE014296 AY119661 AF057167 AAM50315.1 LJIJ01001765 ODM90861.1 CM002912 KMZ01153.1 JH431796 CH902618 EDV41298.1 CH933809 EDW19443.1 CH963847 EDW72607.1 GBBM01000545 JAC34873.1 GEDV01005721 JAP82836.1 CH954178 EDV52749.1 GFPF01006211 MAA17357.1 IACF01000558 LAB66324.1 CH479224 EDW35081.1 CH379070 KRT08893.1 EAL30393.2 LNIX01000021 OXA43806.1 OUUW01000002 SPP76386.1
ODYU01005809 SOQ47068.1 AGBW02007651 OWR54973.1 NEVH01020942 PNF20456.1 KK852686 KDR18468.1 KK107419 EZA51117.1 KZ288269 PBC29884.1 FX985851 BBA93738.1 PNF20457.1 ADTU01003779 GL451091 EFN80062.1 GL888331 EGI62673.1 KQ981768 KYN35998.1 NNAY01001834 OXU22765.1 KQ976701 KYM77235.1 KQ980055 KYN17991.1 GECU01020109 GECU01006522 JAS87597.1 JAT01185.1 GL442175 EFN63841.1 GBYB01008382 JAG78149.1 LJIG01016386 KRT80756.1 KQ977004 KYN06453.1 KQ435765 KOX75351.1 LBMM01003779 KMQ93149.1 AXCN02001905 KQ971354 EFA06143.1 JXUM01141022 JXUM01141023 KQ569173 KXJ68738.1 KQ414613 KOC69059.1 DS235870 EEB19452.1 APCN01005308 AAAB01008968 EAA13221.5 AJWK01028622 GL771866 EFZ09293.1 GFDL01010021 JAV25024.1 CH477243 EAT46335.1 GAMC01017427 JAB89128.1 GEZM01067947 JAV67311.1 KQ759870 OAD62402.1 CVRI01000006 CRK88233.1 GAIX01005073 JAA87487.1 AXCM01002532 CCAG010004106 JXJN01000907 UFQS01002152 UFQT01002152 SSX13347.1 SSX32781.1 GAKP01012197 JAC46755.1 GDHF01004021 JAI48293.1 APGK01028591 APGK01028592 APGK01028593 APGK01028594 KB740694 ENN79436.1 GGFK01008822 MBW42143.1 ADMH02000548 ETN65973.1 ATLV01018954 KE525256 KFB43462.1 GFJQ02007725 JAV99244.1 GGLE01005518 MBY09644.1 PYGN01001881 PSN32349.1 CH940647 EDW70052.2 CH916366 EDV97490.1 CM000159 EDW95512.1 CH480826 EDW44456.1 JRES01000296 KNC32659.1 U34773 AE014296 AY119661 AF057167 AAM50315.1 LJIJ01001765 ODM90861.1 CM002912 KMZ01153.1 JH431796 CH902618 EDV41298.1 CH933809 EDW19443.1 CH963847 EDW72607.1 GBBM01000545 JAC34873.1 GEDV01005721 JAP82836.1 CH954178 EDV52749.1 GFPF01006211 MAA17357.1 IACF01000558 LAB66324.1 CH479224 EDW35081.1 CH379070 KRT08893.1 EAL30393.2 LNIX01000021 OXA43806.1 OUUW01000002 SPP76386.1
Proteomes
UP000053268
UP000283053
UP000218220
UP000007151
UP000235965
UP000027135
+ More
UP000005203 UP000053097 UP000242457 UP000005205 UP000008237 UP000007755 UP000002358 UP000078541 UP000215335 UP000078540 UP000078492 UP000000311 UP000078542 UP000053105 UP000036403 UP000075886 UP000007266 UP000069940 UP000249989 UP000053825 UP000009046 UP000075881 UP000075840 UP000076407 UP000075882 UP000007062 UP000092461 UP000075900 UP000075903 UP000008820 UP000075902 UP000091820 UP000183832 UP000075884 UP000075883 UP000192223 UP000078200 UP000092444 UP000092443 UP000092460 UP000092445 UP000095301 UP000075880 UP000075920 UP000019118 UP000000673 UP000030765 UP000095300 UP000069272 UP000075901 UP000245037 UP000192221 UP000076408 UP000008792 UP000001070 UP000002282 UP000001292 UP000037069 UP000000803 UP000094527 UP000007801 UP000009192 UP000007798 UP000008711 UP000008744 UP000001819 UP000198287 UP000268350
UP000005203 UP000053097 UP000242457 UP000005205 UP000008237 UP000007755 UP000002358 UP000078541 UP000215335 UP000078540 UP000078492 UP000000311 UP000078542 UP000053105 UP000036403 UP000075886 UP000007266 UP000069940 UP000249989 UP000053825 UP000009046 UP000075881 UP000075840 UP000076407 UP000075882 UP000007062 UP000092461 UP000075900 UP000075903 UP000008820 UP000075902 UP000091820 UP000183832 UP000075884 UP000075883 UP000192223 UP000078200 UP000092444 UP000092443 UP000092460 UP000092445 UP000095301 UP000075880 UP000075920 UP000019118 UP000000673 UP000030765 UP000095300 UP000069272 UP000075901 UP000245037 UP000192221 UP000076408 UP000008792 UP000001070 UP000002282 UP000001292 UP000037069 UP000000803 UP000094527 UP000007801 UP000009192 UP000007798 UP000008711 UP000008744 UP000001819 UP000198287 UP000268350
PRIDE
Interpro
CDD
ProteinModelPortal
A0A0N1IKA5
A0A437BIM4
A0A2A4K0C1
A0A2H1W1Y1
A0A212FML7
A0A2J7PVU4
+ More
A0A067RH28 A0A087ZYP9 A0A026W4Y4 A0A2A3EEI5 A0A2Z5U645 A0A2J7PVU0 A0A158NYE9 E2BW30 F4WT60 K7J0U0 A0A195F6A1 A0A232EWQ9 A0A195AYX2 A0A151J561 A0A1B6IKZ6 E2ARV8 A0A0C9R6F3 A0A0T6B0Q4 A0A195D0L0 A0A0N1ITN7 A0A0J7KSE3 A0A182R027 D6WQ19 A0A182G367 A0A0L7RE09 E0W1E6 A0A182K800 A0A182HXI7 A0A182X1S6 A0A182LI74 Q7PMT7 A0A1B0GKA8 E9JD76 A0A182RXG1 A0A182UUN4 A0A1Q3FBR9 Q17I24 W8AWI1 A0A1S4F242 A0A182TVK1 A0A1Y1L3E8 A0A310SMN5 A0A1A9WH97 A0A1J1HPU2 S4PXG6 A0A182NNC6 A0A182M988 A0A1W4XKF0 A0A1A9UP81 A0A1B0FBQ6 A0A1A9XK42 A0A1B0ANP0 A0A1A9ZBX0 A0A336MTA5 A0A1I8M934 A0A034VY24 A0A182IR87 A0A1W4XA81 A0A0K8WAU6 A0A182WCL7 N6UFM6 A0A2M4AMX9 W5JRR2 A0A084VZR8 A0A1I8PRU1 A0A1Z5KW41 A0A182FLR0 A0A2R5LJG8 A0A182SHG1 A0A2P8XK11 A0A1W4UZ38 A0A182YI26 B4LE09 B4IXM8 B4PI64 B4IAZ3 A0A0L0CMG3 Q9VNV3 A0A1D2MD94 A0A0J9S022 T1J2C5 B3MAL1 B4KVA3 B4MN98 A0A023GNS0 A0A131YXC4 B3NJ32 A0A224YI64 A0A2P2HX25 B4H8N8 A0A0R3P8Y6 Q29DM1 A0A226DHY8 A0A3B0JSR4
A0A067RH28 A0A087ZYP9 A0A026W4Y4 A0A2A3EEI5 A0A2Z5U645 A0A2J7PVU0 A0A158NYE9 E2BW30 F4WT60 K7J0U0 A0A195F6A1 A0A232EWQ9 A0A195AYX2 A0A151J561 A0A1B6IKZ6 E2ARV8 A0A0C9R6F3 A0A0T6B0Q4 A0A195D0L0 A0A0N1ITN7 A0A0J7KSE3 A0A182R027 D6WQ19 A0A182G367 A0A0L7RE09 E0W1E6 A0A182K800 A0A182HXI7 A0A182X1S6 A0A182LI74 Q7PMT7 A0A1B0GKA8 E9JD76 A0A182RXG1 A0A182UUN4 A0A1Q3FBR9 Q17I24 W8AWI1 A0A1S4F242 A0A182TVK1 A0A1Y1L3E8 A0A310SMN5 A0A1A9WH97 A0A1J1HPU2 S4PXG6 A0A182NNC6 A0A182M988 A0A1W4XKF0 A0A1A9UP81 A0A1B0FBQ6 A0A1A9XK42 A0A1B0ANP0 A0A1A9ZBX0 A0A336MTA5 A0A1I8M934 A0A034VY24 A0A182IR87 A0A1W4XA81 A0A0K8WAU6 A0A182WCL7 N6UFM6 A0A2M4AMX9 W5JRR2 A0A084VZR8 A0A1I8PRU1 A0A1Z5KW41 A0A182FLR0 A0A2R5LJG8 A0A182SHG1 A0A2P8XK11 A0A1W4UZ38 A0A182YI26 B4LE09 B4IXM8 B4PI64 B4IAZ3 A0A0L0CMG3 Q9VNV3 A0A1D2MD94 A0A0J9S022 T1J2C5 B3MAL1 B4KVA3 B4MN98 A0A023GNS0 A0A131YXC4 B3NJ32 A0A224YI64 A0A2P2HX25 B4H8N8 A0A0R3P8Y6 Q29DM1 A0A226DHY8 A0A3B0JSR4
PDB
4XW3
E-value=9.74198e-51,
Score=508
Ontologies
GO
Topology
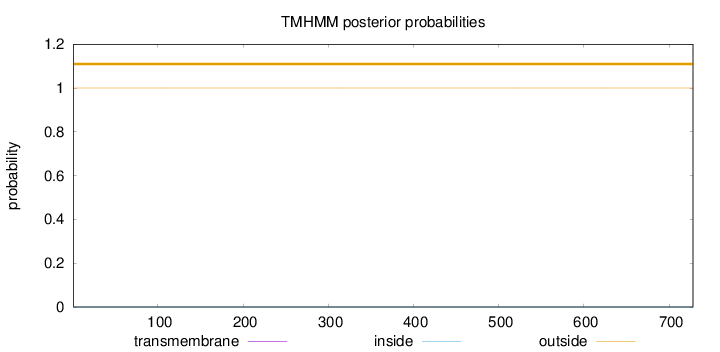
Length:
728
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00241
Exp number, first 60 AAs:
0.00052
Total prob of N-in:
0.00005
outside
1 - 728
Population Genetic Test Statistics
Pi
23.830325
Theta
152.219569
Tajima's D
1.405342
CLR
0.642963
CSRT
0.763161841907905
Interpretation
Uncertain
