Gene
KWMTBOMO09624
Pre Gene Modal
BGIBMGA012853
Annotation
PREDICTED:_histone-lysine_N-methyltransferase_pr-set7_[Amyelois_transitella]
Full name
Histone-lysine N-methyltransferase
+ More
Histone-lysine N-methyltransferase PR-Set7
Histone-lysine N-methyltransferase PR-Set7
Alternative Name
PR/SET domain-containing protein 07
Location in the cell
Cytoplasmic Reliability : 1.79 Mitochondrial Reliability : 1.37 Nuclear Reliability : 1.568
Sequence
CDS
ATGGCAGTTCAAGAAGGGGTAAAGACTCCACATCGCATTGAATTGTGTGAAGACAAGCCTGCTAGGCCCACACGGAATTATAGGAAACGAAGGATTATCACTGCAATACAGCCAAAGACTGAAAATGAAGAACTGAGTTCTGAGCCTCCATTCAAGAAAACTGAACAAAAATCGAAACCATTGAAGACTAGCAACGGTATTTCCAATATGACAAATGGACACTCACTGAGGACACGTGCTGCAAGGGCAGAGATACGTGCAGCGGAAAGCCACAAATTAACAGAGTATTTCAAAGAGGAAATAAAAGTTCCTAAAGTAGAACCTGCTGTATCTCCTTCACCCATAGAAAAGACTGTTAAAGTTGAGAAAGTTTCAAATGGAAATGCAAACCACAAATTAACTGAATTTTTTCCTGTAAGAAGAAGTGTAAGGAAAACATCAAAATGTGTCATGGCTGAGAAAATGAGAGATTTAGAAAGAGCTGTTAGAGAACAAAGAGAAGATGGATTACAAGTTGCCTATTTCGAAGCCAAAGGTCGCGGAGTGATCGCAACCCGTCCGTTTAGTCGCGGTCAGTACGTGGTGGAATATGCCGGTGAACTGATCGGCGTGGCAGAAGCTCGTGAACGAGAACGTTTGTACGCCCTGGATCCTGAGGCCGGATGTTACATGTATTACTTCAGACATGGAGATCAACAGTATTGTATTGATGCAACTGCTGAGTCGGGACGGCTGGGTCGATTGGTGAACCACTCTCGCAACGGTAATCTTCACACAAAGGCGTTGTGGGTGGACGGGCCGCGACTCGTGCTGATCGCTGCACAGGATATCTCCCCGGGGGATGAACTCACTTATGATTATGGAGACCGCTCTAAGGAATCCCTGCAACATCATCCCTGGCTCGCTTTCTGA
Protein
MAVQEGVKTPHRIELCEDKPARPTRNYRKRRIITAIQPKTENEELSSEPPFKKTEQKSKPLKTSNGISNMTNGHSLRTRAARAEIRAAESHKLTEYFKEEIKVPKVEPAVSPSPIEKTVKVEKVSNGNANHKLTEFFPVRRSVRKTSKCVMAEKMRDLERAVREQREDGLQVAYFEAKGRGVIATRPFSRGQYVVEYAGELIGVAEARERERLYALDPEAGCYMYYFRHGDQQYCIDATAESGRLGRLVNHSRNGNLHTKALWVDGPRLVLIAAQDISPGDELTYDYGDRSKESLQHHPWLAF
Summary
Description
Histone methyltransferase that specifically monomethylates 'Lys-20' of histone H4. H4 'Lys-20' monomethylation is enriched during mitosis and represents a specific tag for epigenetic transcriptional repression. Mainly functions in euchromatin regions, thereby playing a central role in the silencing of euchromatic genes. Required for cell proliferation, possibly by contributing to the maintenance of proper higher-order structure of DNA and chromosome condensation during mitosis (By similarity).
Catalytic Activity
L-lysyl-[histone] + S-adenosyl-L-methionine = H(+) + N(6)-methyl-L-lysyl-[histone] + S-adenosyl-L-homocysteine
Similarity
Belongs to the class V-like SAM-binding methyltransferase superfamily. Histone-lysine methyltransferase family. PR/SET subfamily.
Keywords
Cell cycle
Cell division
Chromatin regulator
Chromosome
Complete proteome
Methyltransferase
Mitosis
Nucleus
Reference proteome
Repressor
S-adenosyl-L-methionine
Transcription
Transcription regulation
Transferase
Feature
chain Histone-lysine N-methyltransferase PR-Set7
Uniprot
EC Number
2.1.1.43
EMBL
BABH01003712
NWSH01000320
PCG77498.1
ODYU01000646
SOQ35769.1
RSAL01000051
+ More
RVE50291.1 AGBW02007651 OWR54974.1 LADI01006439 KPJ20645.1 KQ460208 KPJ16612.1 GDQN01011257 JAT79797.1 PCG77497.1 JTDY01000139 KOB78637.1 GANO01003982 JAB55889.1 KK854023 PTY09592.1 GDIP01144006 LRGB01000930 JAJ79396.1 KZS15065.1 GDIQ01167221 JAK84504.1 CM000070 GDIP01067060 JAM36655.1 GDIQ01017496 JAN77241.1 GDIQ01102654 JAL49072.1
RVE50291.1 AGBW02007651 OWR54974.1 LADI01006439 KPJ20645.1 KQ460208 KPJ16612.1 GDQN01011257 JAT79797.1 PCG77497.1 JTDY01000139 KOB78637.1 GANO01003982 JAB55889.1 KK854023 PTY09592.1 GDIP01144006 LRGB01000930 JAJ79396.1 KZS15065.1 GDIQ01167221 JAK84504.1 CM000070 GDIP01067060 JAM36655.1 GDIQ01017496 JAN77241.1 GDIQ01102654 JAL49072.1
Proteomes
Pfam
PF00856 SET
ProteinModelPortal
PDB
5HQ2
E-value=1.3817e-48,
Score=486
Ontologies
GO
Topology
Subcellular location
Nucleus
Specifically localizes to mitotic chromosomes. Associates to chromatin-dense and transcriptionally silent euchromatic regions (By similarity). With evidence from 1 publications.
Chromosome Specifically localizes to mitotic chromosomes. Associates to chromatin-dense and transcriptionally silent euchromatic regions (By similarity). With evidence from 1 publications.
Chromosome Specifically localizes to mitotic chromosomes. Associates to chromatin-dense and transcriptionally silent euchromatic regions (By similarity). With evidence from 1 publications.
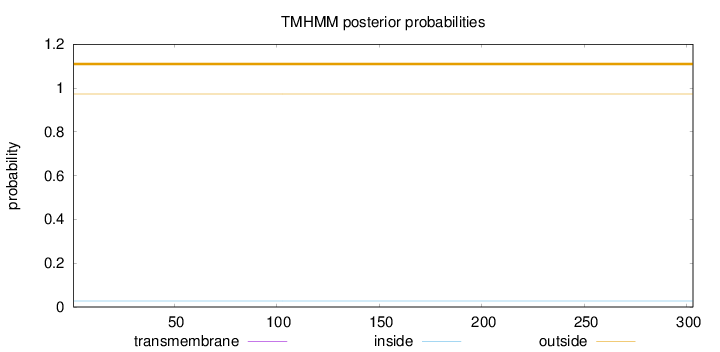
Length:
303
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02741
outside
1 - 303
Population Genetic Test Statistics
Pi
173.353584
Theta
170.423143
Tajima's D
0.140835
CLR
0.409033
CSRT
0.408829558522074
Interpretation
Uncertain
