Gene
KWMTBOMO09623
Pre Gene Modal
BGIBMGA012984
Annotation
PREDICTED:_histone_deacetylase_8-like_[Bombyx_mori]
Full name
Histone deacetylase
+ More
Histone deacetylase 8
Histone deacetylase 8
Location in the cell
Cytoplasmic Reliability : 1.649
Sequence
CDS
ATGAACAATGCTCGTGTTGCTTATCTTTGGGATGAGAAATTAGTGAAGGAATGTATTAGGCTACCAGCAGTATTCGGCAGGGCTCGACTCGTGCACAATTTAATCGAAGCATATGGTCTGATTAGTAAATTGAAAGTAATCCGTTCATCTCCAGCAAGTTATGAAGATTTAAACGTTTTTCATTCAGACTTATATTTAGAACATTTAAAACAAATAACAGACATTGATGATGATTACATTTCTAATGCTCAAGATGAAAATTTTGGGATAGGATATGATTGTCCCCCTGTCCCTAATATGTTCGAGCTGGTGTCAACCATTGCTGGAGGTTCAGTGACGGCTGCTAAGTGTTTGACGATGGGTATTGCTGATATAGCCATTAATTGGTGTGGGGGCTGGCATCATGCACATAATAATCGTGCAGAGGGTTTCTGTTACGTAAACGACATTGTAATAGCAATAGAAAAATTGAAAGGAAAATTCAAAAATATTTTATACGTTGATTTAGATGTGCATCATGGCAATGGTGTTCAAGATGCTTATTGGACGACTCGGTCAGTATACACATTGTCTTTTCACAAGTTCGAACCAGGTTTCTATCCAGGTACAGGGAGTATAGAAGACATTGGTTGCGGTGACGGTGAAGGATATTCATGTAACTTCCCACTTAATGAAGCTTATTCCGATAATACAATTCAATTTGTTTTTGAAAAGGTATTCAATTCTGTTTATTCGAATTTTGCTCCTGACGCAATCGTAGTACAATGCGGGGCTGACGCATTGTCTCATGATCCGCACGGCGGTGCTTGTTTGACTAGTCGCGGTTATTGCGCCTGCATAAGCACTGTTCTCGATAAACAAAAACCTACAATGCTACTCGGAGGGGGAGGATACAACCACTCAAATACTGCTCGTCTGTGGACAACCTTGACAGCTTTCGTTACTGGTGTAGAATTAGATGAGATAATTCCTGAACACAATGAGTGGCCTGAGTATGGACCAGATTACATGCTTCCTGTTCAAGGCACCCTTATTAAGGATACAAACAAGAAATCTTATCTTGAAGACTGTATAAATAAAATAAATGAAAACTTGAAAAAACACTTAGCCAAAACGGAAAAACTTGAACCTGAATCAAAAATATATAAAACAGGTGGAGAATTGGTAAAAAGTGATTATAAAATAAACAAGAGCTATATTCCTAAATATAGTTATGTGCAGAAGACTGAAAGTACCTGTGATACAATGCAGCCTGTTGATGTATACGACTTTGTAGAATAA
Protein
MNNARVAYLWDEKLVKECIRLPAVFGRARLVHNLIEAYGLISKLKVIRSSPASYEDLNVFHSDLYLEHLKQITDIDDDYISNAQDENFGIGYDCPPVPNMFELVSTIAGGSVTAAKCLTMGIADIAINWCGGWHHAHNNRAEGFCYVNDIVIAIEKLKGKFKNILYVDLDVHHGNGVQDAYWTTRSVYTLSFHKFEPGFYPGTGSIEDIGCGDGEGYSCNFPLNEAYSDNTIQFVFEKVFNSVYSNFAPDAIVVQCGADALSHDPHGGACLTSRGYCACISTVLDKQKPTMLLGGGGYNHSNTARLWTTLTAFVTGVELDEIIPEHNEWPEYGPDYMLPVQGTLIKDTNKKSYLEDCINKINENLKKHLAKTEKLEPESKIYKTGGELVKSDYKINKSYIPKYSYVQKTESTCDTMQPVDVYDFVE
Summary
Description
Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. Histone deacetylases act via the formation of large multiprotein complexes. Also involved in the deacetylation of cohesin complex protein SMC3 regulating release of cohesin complexes from chromatin. May play a role in smooth muscle cell contractility.
Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. Histone deacetylases act via the formation of large multiprotein complexes. Also involved in the deacetylation of cohesin complex protein SMC3 regulating release of cohesin complexes from chromatin. May play a role in smooth muscle cell contractility (By similarity).
Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. Histone deacetylases act via the formation of large multiprotein complexes. Also involved in the deacetylation of cohesin complex protein SMC3 regulating release of cohesin complexes from chromatin. May play a role in smooth muscle cell contractility (By similarity).
Catalytic Activity
Hydrolysis of an N(6)-acetyl-lysine residue of a histone to yield a deacetylated histone.
Cofactor
a divalent metal cation
Subunit
Interacts with PEPB2-MYH11, a fusion protein consisting of the 165 N-terminal residues of CBF-beta (PEPB2) with the tail region of MYH11 produced by the inversion Inv(16)(p13q22), a translocation associated with acute myeloid leukemia of M4EO subtype. The PEPB2-MYH1 fusion protein also interacts with RUNX1, a well known transcriptional regulator, suggesting that the interaction with HDAC8 may participate in the conversion of RUNX1 into a constitutive transcriptional repressor. Interacts with CBFA2T3. Interacts with phosphorylated SMG5/EST1B; this interaction protects SMG5 from ubiquitin-mediated degradation. Associates with alpha-SMA (smooth muscle alpha-actin).
Interacts with CBFA2T3. Interacts with phosphorylated SMG5/EST1B; this interaction protects SMG5 from ubiquitin-mediated degradation. Associates with alpha-SMA (smooth muscle alpha-actin) (By similarity).
Interacts with CBFA2T3. Interacts with phosphorylated SMG5/EST1B; this interaction protects SMG5 from ubiquitin-mediated degradation. Associates with alpha-SMA (smooth muscle alpha-actin) (By similarity).
Similarity
Belongs to the histone deacetylase family. HD Type 1 subfamily.
Belongs to the histone deacetylase family. HD type 1 subfamily.
Belongs to the histone deacetylase family. HD type 1 subfamily.
Keywords
3D-structure
Alternative splicing
Chromatin regulator
Complete proteome
Cytoplasm
Disease mutation
Hydrolase
Mental retardation
Metal-binding
Nucleus
Obesity
Phosphoprotein
Reference proteome
Repressor
Transcription
Transcription regulation
Feature
chain Histone deacetylase 8
splice variant In isoform 4 and isoform 6.
sequence variant In CDLS5; dbSNP:rs397515416.
splice variant In isoform 4 and isoform 6.
sequence variant In CDLS5; dbSNP:rs397515416.
Uniprot
A0A2A4JZU2
A0A2H1V4J4
A0A437BIM0
A0A212FMK1
A0A0N0PEY2
A0A2P8YTB3
+ More
A0A0P4WDA7 A0A2S2PC45 A0A0A9XY16 A0A401NUI0 J9K941 A0A0P4XN91 A0A0P6EZ59 A0A067R8A7 A0A0P5TRJ8 A0A0P5VY96 A0A0N8BLN4 A0A0N8EN19 A0A2H8TV52 X1X3E1 A0A444SYB5 A0A0P6H279 A0A401S1S7 A0A0N8AAC4 A0A0P5Y1K8 A0A0P4Z3G1 A0A369S5W8 A0A0P6AJT9 A0A023F1K9 A0A0P4Z0X0 A0A0Q3URX1 A0A3B4URL5 V9KWM5 B3RYK4 A0A218V4F9 A0A336MDH4 A0A1Y1M2X5 H0Z315 F6SGB2 H9ESD4 A0A0P5J6E7 A0A336MM84 A0A069DT17 G7Q312 A0A1D5QWF8 F1NFY6 A0A2I0TNH8 A0A151P891 C1BMT8 A0A1B6I328 A0A2J7PYA2 W5NAX4 A0A2K6C7N2 H3ATC2 U3FL58 H9GLC3 A0A1A8P3V1 A0A3P8UKT1 A0A2K6ELQ3 A0A1S2ZXI8 A0A0P5VKV5 A0A1A8HDA1 A0A1A8JPV9 A0A1A8VFK7 A0A2R8ZNP2 A0A2I3TSB3 Q9BY41 A0A2T7P3C8 A0A3Q2CY37 A0A2K5QLL7 U3KBX2 A0A2K5H932 G3R7G9 A0A2J8R852 K9ITE1 A0A2I0LM71 A0A2K6TG81 G1LGX4 A0A2K5DZM8 A0A3B4DQH3 A0A0F7ZDP5 T1E5I7 A0A1P8L011 A0A1A8BMQ3 R7UB93 G3WQV6 A0A1S3HC34 F6TCX9 K7B694 A0A146NLK8 A0A1A8LJR4 U6CNQ8 G1T563 A0A3Q3LQH6 A0A0P6J8M3 A0A2Y9DTB3 C3ZM32 I3MBM8 S7PQ76 B1WC68
A0A0P4WDA7 A0A2S2PC45 A0A0A9XY16 A0A401NUI0 J9K941 A0A0P4XN91 A0A0P6EZ59 A0A067R8A7 A0A0P5TRJ8 A0A0P5VY96 A0A0N8BLN4 A0A0N8EN19 A0A2H8TV52 X1X3E1 A0A444SYB5 A0A0P6H279 A0A401S1S7 A0A0N8AAC4 A0A0P5Y1K8 A0A0P4Z3G1 A0A369S5W8 A0A0P6AJT9 A0A023F1K9 A0A0P4Z0X0 A0A0Q3URX1 A0A3B4URL5 V9KWM5 B3RYK4 A0A218V4F9 A0A336MDH4 A0A1Y1M2X5 H0Z315 F6SGB2 H9ESD4 A0A0P5J6E7 A0A336MM84 A0A069DT17 G7Q312 A0A1D5QWF8 F1NFY6 A0A2I0TNH8 A0A151P891 C1BMT8 A0A1B6I328 A0A2J7PYA2 W5NAX4 A0A2K6C7N2 H3ATC2 U3FL58 H9GLC3 A0A1A8P3V1 A0A3P8UKT1 A0A2K6ELQ3 A0A1S2ZXI8 A0A0P5VKV5 A0A1A8HDA1 A0A1A8JPV9 A0A1A8VFK7 A0A2R8ZNP2 A0A2I3TSB3 Q9BY41 A0A2T7P3C8 A0A3Q2CY37 A0A2K5QLL7 U3KBX2 A0A2K5H932 G3R7G9 A0A2J8R852 K9ITE1 A0A2I0LM71 A0A2K6TG81 G1LGX4 A0A2K5DZM8 A0A3B4DQH3 A0A0F7ZDP5 T1E5I7 A0A1P8L011 A0A1A8BMQ3 R7UB93 G3WQV6 A0A1S3HC34 F6TCX9 K7B694 A0A146NLK8 A0A1A8LJR4 U6CNQ8 G1T563 A0A3Q3LQH6 A0A0P6J8M3 A0A2Y9DTB3 C3ZM32 I3MBM8 S7PQ76 B1WC68
EC Number
3.5.1.98
Pubmed
22118469
26354079
29403074
25401762
26823975
30297745
+ More
24845553 30042472 25474469 24402279 18719581 28004739 20360741 17495919 25319552 26334808 22002653 17431167 15592404 22293439 9215903 25243066 21881562 24487278 22722832 16136131 10748112 10926844 10922473 8889548 14702039 15772651 15489334 10756090 11533236 12509458 14701748 15772115 16538051 16809764 22889856 22885700 15242608 17721440 19053282 22398555 23371554 20010809 23758969 25727380 23254933 21709235 21993624 18563158
24845553 30042472 25474469 24402279 18719581 28004739 20360741 17495919 25319552 26334808 22002653 17431167 15592404 22293439 9215903 25243066 21881562 24487278 22722832 16136131 10748112 10926844 10922473 8889548 14702039 15772651 15489334 10756090 11533236 12509458 14701748 15772115 16538051 16809764 22889856 22885700 15242608 17721440 19053282 22398555 23371554 20010809 23758969 25727380 23254933 21709235 21993624 18563158
EMBL
NWSH01000320
PCG77507.1
ODYU01000646
SOQ35770.1
RSAL01000051
RVE50290.1
+ More
AGBW02007651 OWR54975.1 LADI01006439 KPJ20644.1 PYGN01000374 PSN47474.1 GDRN01072091 JAI63597.1 GGMR01014418 MBY27037.1 GBHO01018780 GBRD01009922 GDHC01020413 JAG24824.1 JAG55902.1 JAP98215.1 BFAA01004108 GCB64500.1 ABLF02031468 ABLF02031473 GDIP01241527 LRGB01000944 JAI81874.1 KZS14791.1 GDIQ01057221 GDIQ01033362 JAN37516.1 KK852673 KDR18741.1 GDIP01124908 JAL78806.1 GDIP01093509 JAM10206.1 GDIQ01165587 JAK86138.1 GDIQ01010716 JAN84021.1 GFXV01006015 MBW17820.1 ABLF02034889 SAUD01018877 RXG58395.1 GDIQ01035880 JAN58857.1 BEZZ01000055 GCC24345.1 GDIP01166945 JAJ56457.1 GDIP01064267 JAM39448.1 GDIP01221139 JAJ02263.1 NOWV01000057 RDD42307.1 GDIP01028696 JAM75019.1 GBBI01003362 JAC15350.1 GDIP01222336 JAJ01066.1 LMAW01002620 KQK78994.1 JW870226 AFP02744.1 DS985245 EDV25056.1 MUZQ01000056 OWK60621.1 UFQS01000750 UFQT01000750 SSX06612.1 SSX26959.1 GEZM01043555 JAV79030.1 ABQF01018743 JU321537 JU476398 JV047641 AFE65293.1 AFH33202.1 AFI37712.1 GDIQ01202514 JAK49211.1 UFQT01001504 SSX30855.1 GBGD01002043 JAC86846.1 AQIA01085334 AQIA01085335 AQIA01085336 AQIA01085337 AQIA01085338 AQIA01085339 AQIA01085340 AQIA01085341 CM001296 EHH60991.1 JSUE03045885 JSUE03045886 JSUE03045887 CM001273 EHH30843.1 AADN05000020 KZ508327 PKU35340.1 AKHW03000635 KYO45119.1 BT075917 ACO10341.1 GECU01026431 JAS81275.1 NEVH01020850 PNF21315.1 AHAT01015581 AHAT01015582 AHAT01015583 AHAT01015584 AFYH01043253 AFYH01043254 AFYH01043255 AFYH01043256 GAMT01008549 GAMS01009942 GAMR01005256 GAMQ01006531 GAMP01008046 GAMP01008045 GAMP01008044 JAB03312.1 JAB13194.1 JAB28676.1 JAB35320.1 JAB44711.1 AAWZ02031887 HAEI01001388 HAEH01010318 SBR75961.1 GDIP01098602 JAM05113.1 HAEB01008146 HAEC01013236 SBQ81453.1 HAED01011632 HAEE01002128 SBR22148.1 HADY01002754 HAEJ01017910 SBS58367.1 AJFE02013641 AJFE02013642 AJFE02013643 AJFE02013644 AJFE02013645 AJFE02013646 AJFE02013647 AACZ04026470 AACZ04026471 AACZ04026472 AACZ04026473 AACZ04026474 NBAG03000573 PNI14692.1 AF230097 AF245664 AJ277724 BQ189619 AK296641 AK300895 AA376331 AI159768 T99283 AF212246 AL133500 BX295542 BC050433 PZQS01000006 PVD27935.1 AGTO01001454 CABD030125736 CABD030125737 CABD030125738 CABD030125739 CABD030125740 CABD030125741 CABD030125742 CABD030125743 CABD030125744 CABD030125745 NDHI03003732 PNJ04702.1 GABZ01001828 JAA51697.1 AKCR02000199 PKK18529.1 ACTA01034718 ACTA01042717 ACTA01050717 ACTA01058717 ACTA01066717 ACTA01074717 ACTA01082717 ACTA01090717 GBEX01001531 JAI13029.1 GAAZ01001085 GBKC01001614 JAA96858.1 JAG44456.1 KY273094 APW77295.1 HADZ01004513 SBP68454.1 AMQN01001497 KB303020 ELU03635.1 AEFK01131716 GABC01006898 GABF01006427 GABD01007946 GABE01010319 JAA04440.1 JAA15718.1 JAA25154.1 JAA34420.1 GCES01153599 JAQ32723.1 HAEF01007189 HAEG01007292 SBR44571.1 HAAF01000265 CCP72091.1 AAGW02057537 AAGW02057538 AAGW02057539 AAGW02057540 AAGW02057541 AAGW02057542 AAGW02057543 AAGW02057544 AAGW02057545 AAGW02057546 GEBF01003730 JAN99902.1 GG666644 EEN46288.1 AGTP01104297 AGTP01104298 AGTP01104299 AGTP01104300 AGTP01104301 AGTP01104302 AGTP01104303 AGTP01104304 KE163658 EPQ13013.1 BC162023
AGBW02007651 OWR54975.1 LADI01006439 KPJ20644.1 PYGN01000374 PSN47474.1 GDRN01072091 JAI63597.1 GGMR01014418 MBY27037.1 GBHO01018780 GBRD01009922 GDHC01020413 JAG24824.1 JAG55902.1 JAP98215.1 BFAA01004108 GCB64500.1 ABLF02031468 ABLF02031473 GDIP01241527 LRGB01000944 JAI81874.1 KZS14791.1 GDIQ01057221 GDIQ01033362 JAN37516.1 KK852673 KDR18741.1 GDIP01124908 JAL78806.1 GDIP01093509 JAM10206.1 GDIQ01165587 JAK86138.1 GDIQ01010716 JAN84021.1 GFXV01006015 MBW17820.1 ABLF02034889 SAUD01018877 RXG58395.1 GDIQ01035880 JAN58857.1 BEZZ01000055 GCC24345.1 GDIP01166945 JAJ56457.1 GDIP01064267 JAM39448.1 GDIP01221139 JAJ02263.1 NOWV01000057 RDD42307.1 GDIP01028696 JAM75019.1 GBBI01003362 JAC15350.1 GDIP01222336 JAJ01066.1 LMAW01002620 KQK78994.1 JW870226 AFP02744.1 DS985245 EDV25056.1 MUZQ01000056 OWK60621.1 UFQS01000750 UFQT01000750 SSX06612.1 SSX26959.1 GEZM01043555 JAV79030.1 ABQF01018743 JU321537 JU476398 JV047641 AFE65293.1 AFH33202.1 AFI37712.1 GDIQ01202514 JAK49211.1 UFQT01001504 SSX30855.1 GBGD01002043 JAC86846.1 AQIA01085334 AQIA01085335 AQIA01085336 AQIA01085337 AQIA01085338 AQIA01085339 AQIA01085340 AQIA01085341 CM001296 EHH60991.1 JSUE03045885 JSUE03045886 JSUE03045887 CM001273 EHH30843.1 AADN05000020 KZ508327 PKU35340.1 AKHW03000635 KYO45119.1 BT075917 ACO10341.1 GECU01026431 JAS81275.1 NEVH01020850 PNF21315.1 AHAT01015581 AHAT01015582 AHAT01015583 AHAT01015584 AFYH01043253 AFYH01043254 AFYH01043255 AFYH01043256 GAMT01008549 GAMS01009942 GAMR01005256 GAMQ01006531 GAMP01008046 GAMP01008045 GAMP01008044 JAB03312.1 JAB13194.1 JAB28676.1 JAB35320.1 JAB44711.1 AAWZ02031887 HAEI01001388 HAEH01010318 SBR75961.1 GDIP01098602 JAM05113.1 HAEB01008146 HAEC01013236 SBQ81453.1 HAED01011632 HAEE01002128 SBR22148.1 HADY01002754 HAEJ01017910 SBS58367.1 AJFE02013641 AJFE02013642 AJFE02013643 AJFE02013644 AJFE02013645 AJFE02013646 AJFE02013647 AACZ04026470 AACZ04026471 AACZ04026472 AACZ04026473 AACZ04026474 NBAG03000573 PNI14692.1 AF230097 AF245664 AJ277724 BQ189619 AK296641 AK300895 AA376331 AI159768 T99283 AF212246 AL133500 BX295542 BC050433 PZQS01000006 PVD27935.1 AGTO01001454 CABD030125736 CABD030125737 CABD030125738 CABD030125739 CABD030125740 CABD030125741 CABD030125742 CABD030125743 CABD030125744 CABD030125745 NDHI03003732 PNJ04702.1 GABZ01001828 JAA51697.1 AKCR02000199 PKK18529.1 ACTA01034718 ACTA01042717 ACTA01050717 ACTA01058717 ACTA01066717 ACTA01074717 ACTA01082717 ACTA01090717 GBEX01001531 JAI13029.1 GAAZ01001085 GBKC01001614 JAA96858.1 JAG44456.1 KY273094 APW77295.1 HADZ01004513 SBP68454.1 AMQN01001497 KB303020 ELU03635.1 AEFK01131716 GABC01006898 GABF01006427 GABD01007946 GABE01010319 JAA04440.1 JAA15718.1 JAA25154.1 JAA34420.1 GCES01153599 JAQ32723.1 HAEF01007189 HAEG01007292 SBR44571.1 HAAF01000265 CCP72091.1 AAGW02057537 AAGW02057538 AAGW02057539 AAGW02057540 AAGW02057541 AAGW02057542 AAGW02057543 AAGW02057544 AAGW02057545 AAGW02057546 GEBF01003730 JAN99902.1 GG666644 EEN46288.1 AGTP01104297 AGTP01104298 AGTP01104299 AGTP01104300 AGTP01104301 AGTP01104302 AGTP01104303 AGTP01104304 KE163658 EPQ13013.1 BC162023
Proteomes
UP000218220
UP000283053
UP000007151
UP000053268
UP000245037
UP000288216
+ More
UP000007819 UP000076858 UP000027135 UP000288706 UP000287033 UP000253843 UP000051836 UP000261420 UP000009022 UP000197619 UP000007754 UP000002280 UP000009130 UP000233100 UP000006718 UP000000539 UP000050525 UP000235965 UP000018468 UP000233120 UP000008672 UP000001646 UP000265120 UP000233160 UP000079721 UP000240080 UP000002277 UP000005640 UP000245119 UP000265020 UP000233040 UP000016665 UP000233080 UP000001519 UP000053872 UP000233220 UP000008912 UP000233020 UP000261440 UP000014760 UP000007648 UP000085678 UP000008225 UP000265000 UP000001811 UP000261640 UP000248480 UP000001554 UP000005215 UP000002494
UP000007819 UP000076858 UP000027135 UP000288706 UP000287033 UP000253843 UP000051836 UP000261420 UP000009022 UP000197619 UP000007754 UP000002280 UP000009130 UP000233100 UP000006718 UP000000539 UP000050525 UP000235965 UP000018468 UP000233120 UP000008672 UP000001646 UP000265120 UP000233160 UP000079721 UP000240080 UP000002277 UP000005640 UP000245119 UP000265020 UP000233040 UP000016665 UP000233080 UP000001519 UP000053872 UP000233220 UP000008912 UP000233020 UP000261440 UP000014760 UP000007648 UP000085678 UP000008225 UP000265000 UP000001811 UP000261640 UP000248480 UP000001554 UP000005215 UP000002494
Pfam
PF00850 Hist_deacetyl
Interpro
SUPFAM
SSF52768
SSF52768
Gene 3D
ProteinModelPortal
A0A2A4JZU2
A0A2H1V4J4
A0A437BIM0
A0A212FMK1
A0A0N0PEY2
A0A2P8YTB3
+ More
A0A0P4WDA7 A0A2S2PC45 A0A0A9XY16 A0A401NUI0 J9K941 A0A0P4XN91 A0A0P6EZ59 A0A067R8A7 A0A0P5TRJ8 A0A0P5VY96 A0A0N8BLN4 A0A0N8EN19 A0A2H8TV52 X1X3E1 A0A444SYB5 A0A0P6H279 A0A401S1S7 A0A0N8AAC4 A0A0P5Y1K8 A0A0P4Z3G1 A0A369S5W8 A0A0P6AJT9 A0A023F1K9 A0A0P4Z0X0 A0A0Q3URX1 A0A3B4URL5 V9KWM5 B3RYK4 A0A218V4F9 A0A336MDH4 A0A1Y1M2X5 H0Z315 F6SGB2 H9ESD4 A0A0P5J6E7 A0A336MM84 A0A069DT17 G7Q312 A0A1D5QWF8 F1NFY6 A0A2I0TNH8 A0A151P891 C1BMT8 A0A1B6I328 A0A2J7PYA2 W5NAX4 A0A2K6C7N2 H3ATC2 U3FL58 H9GLC3 A0A1A8P3V1 A0A3P8UKT1 A0A2K6ELQ3 A0A1S2ZXI8 A0A0P5VKV5 A0A1A8HDA1 A0A1A8JPV9 A0A1A8VFK7 A0A2R8ZNP2 A0A2I3TSB3 Q9BY41 A0A2T7P3C8 A0A3Q2CY37 A0A2K5QLL7 U3KBX2 A0A2K5H932 G3R7G9 A0A2J8R852 K9ITE1 A0A2I0LM71 A0A2K6TG81 G1LGX4 A0A2K5DZM8 A0A3B4DQH3 A0A0F7ZDP5 T1E5I7 A0A1P8L011 A0A1A8BMQ3 R7UB93 G3WQV6 A0A1S3HC34 F6TCX9 K7B694 A0A146NLK8 A0A1A8LJR4 U6CNQ8 G1T563 A0A3Q3LQH6 A0A0P6J8M3 A0A2Y9DTB3 C3ZM32 I3MBM8 S7PQ76 B1WC68
A0A0P4WDA7 A0A2S2PC45 A0A0A9XY16 A0A401NUI0 J9K941 A0A0P4XN91 A0A0P6EZ59 A0A067R8A7 A0A0P5TRJ8 A0A0P5VY96 A0A0N8BLN4 A0A0N8EN19 A0A2H8TV52 X1X3E1 A0A444SYB5 A0A0P6H279 A0A401S1S7 A0A0N8AAC4 A0A0P5Y1K8 A0A0P4Z3G1 A0A369S5W8 A0A0P6AJT9 A0A023F1K9 A0A0P4Z0X0 A0A0Q3URX1 A0A3B4URL5 V9KWM5 B3RYK4 A0A218V4F9 A0A336MDH4 A0A1Y1M2X5 H0Z315 F6SGB2 H9ESD4 A0A0P5J6E7 A0A336MM84 A0A069DT17 G7Q312 A0A1D5QWF8 F1NFY6 A0A2I0TNH8 A0A151P891 C1BMT8 A0A1B6I328 A0A2J7PYA2 W5NAX4 A0A2K6C7N2 H3ATC2 U3FL58 H9GLC3 A0A1A8P3V1 A0A3P8UKT1 A0A2K6ELQ3 A0A1S2ZXI8 A0A0P5VKV5 A0A1A8HDA1 A0A1A8JPV9 A0A1A8VFK7 A0A2R8ZNP2 A0A2I3TSB3 Q9BY41 A0A2T7P3C8 A0A3Q2CY37 A0A2K5QLL7 U3KBX2 A0A2K5H932 G3R7G9 A0A2J8R852 K9ITE1 A0A2I0LM71 A0A2K6TG81 G1LGX4 A0A2K5DZM8 A0A3B4DQH3 A0A0F7ZDP5 T1E5I7 A0A1P8L011 A0A1A8BMQ3 R7UB93 G3WQV6 A0A1S3HC34 F6TCX9 K7B694 A0A146NLK8 A0A1A8LJR4 U6CNQ8 G1T563 A0A3Q3LQH6 A0A0P6J8M3 A0A2Y9DTB3 C3ZM32 I3MBM8 S7PQ76 B1WC68
PDB
5D1D
E-value=2.05336e-90,
Score=848
Ontologies
GO
GO:0032041
GO:0046872
GO:0005634
GO:0004407
GO:0016021
GO:0045944
GO:0070933
GO:0070932
GO:0000118
GO:0031647
GO:0000228
GO:0006333
GO:0000122
GO:0031397
GO:0008134
GO:0005886
GO:0006325
GO:0051879
GO:0007062
GO:0071922
GO:0032204
GO:0030544
GO:0005737
GO:0005654
GO:0035984
GO:0010629
GO:0019213
GO:0003682
GO:2000616
GO:0045668
GO:1904322
GO:0016575
GO:0005852
GO:0006413
GO:0031369
GO:0003723
GO:0006730
GO:0009258
GO:0016155
GO:0003824
Topology
Subcellular location
Nucleus
Cytoplasm
Cytoplasm
Length:
426
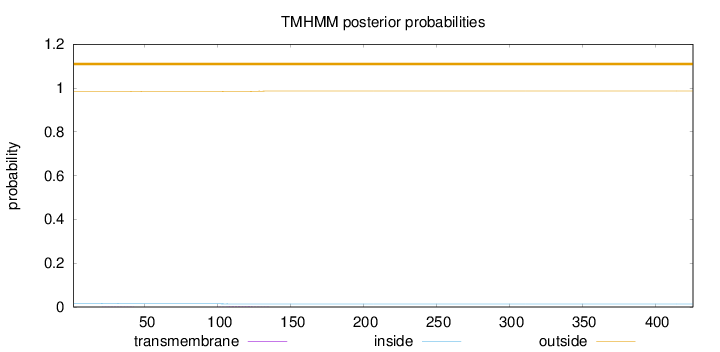
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0456600000000001
Exp number, first 60 AAs:
0.00647
Total prob of N-in:
0.01550
outside
1 - 426
Population Genetic Test Statistics
Pi
145.821776
Theta
40.368953
Tajima's D
-2.313878
CLR
1592.771279
CSRT
0.00269986500674966
Interpretation
Possibly Positive selection
