Gene
KWMTBOMO09617
Annotation
PREDICTED:_pickpocket_protein_28-like_[Bombyx_mori]
Full name
Pickpocket protein 28
Location in the cell
Extracellular Reliability : 1.069 Mitochondrial Reliability : 1.065 PlasmaMembrane Reliability : 1.107
Sequence
CDS
ATGCTGGTAGTGCACTTTTTCTTTGAAGACAACACCTTCTTGAGGTTCACAAAGGGTGAAATATTTGGACTAACAGAATTTTTATCGAACACGGGTGGCCTTCTAGGATTGTGTATGGGCTTCAGTATTATGAGTGCCGTAGAACTCATATACTACCTCACTCTCCGAGCGCTGTGCATGGCAAGACGACGTCCAAAGCATCCTTTCACTAATTAA
Protein
MLVVHFFFEDNTFLRFTKGEIFGLTEFLSNTGGLLGLCMGFSIMSAVELIYYLTLRALCMARRRPKHPFTN
Summary
Description
Osmosensitive ion channel that mediates the cellular and behavioral response to water. Plays an essential role in gustatory water reception. Part of a complex that plays a role in tracheal liquid clearance. Probable role in sodium transport.
Similarity
Belongs to the amiloride-sensitive sodium channel (TC 1.A.6) family.
Keywords
Cell membrane
Complete proteome
Ion channel
Ion transport
Membrane
Reference proteome
Sodium
Sodium channel
Sodium transport
Transmembrane
Transmembrane helix
Transport
Feature
chain Pickpocket protein 28
Uniprot
A0A2A4JZR3
A0A437BIK4
A0A2H1V4S6
A0A026WH41
A0A3L8DZA0
A0A1I8NLF6
+ More
A0A1B0ARX1 A0A1I8MB58 A0A1I8M676 B4NE18 A0A1A9YCN5 B4L1U3 A0A0P8Y7G7 B3N0C8 A0A1B0FNM8 A0A0L0C993 A0A0M9A038 A0A3B0J8E7 A0A1J1IXR0 Q29IB8 A0A1A9UHP7 B4M1J5 B4JL61 A0A2S2NS56 A0A0M5J642 A0A154PLS7 A0A1W4VHZ0 B4Q1W1 B3NW89 B4IEU0 A0A2J7PMQ8 B4R639 Q86LG1 A0A1B0GN28 A0A1B6MGD0 A0A158NBF0 A0A067RB77 A0A0C9RL33 A0A1S4EW92 A0A1A9WF90 A0A1D2MTK5 A0A232FIR5 B0WIZ9 K7IP12 A0A1D2N8C1 A0A226EXG1 A0A1I8N2I2 A0A1W4XR61 A0A1I8MQ88 A0A2W1BGB8 A0A139WCM4 B0WDC8 A0A226DYB2 A0A182GC76 B0WDC6 D6WXM7 A0A1D2MEU0 A0A226EYV0 A0A3S2NPU4 Q17NS9 A0A194PW83 A0A226F6M1 A0A1I8PEH0 A0A182MXN6 A0A1J1HMY9 A0A0L7L5Y6 A0A1Y1L465 Q17MX3 B0X0S1 A0A1I8PI06 N6U8E9 A0A1S4EX90 A0A182GRH0 T1H366 U4UGG3 Q9VS73 B4PC79 B3NFC4 A0A0J9RQW9 B4HV75 A0A182H7C8 A0A084WQC5 A0A1I8NHK4 B0WKI1 A0A1B0GL64 A0A226F2X9 A0A1S4G1A5 A0A226EYJ2 W5JCG6 B4J292 B3M5Z2 Q2M133 B4GUR0 A0A1B0GH50 A0A0L7LLW2 A0A0Q9WPD7 D6WXM6 A0A1I8N4A3 A0A182WEP1
A0A1B0ARX1 A0A1I8MB58 A0A1I8M676 B4NE18 A0A1A9YCN5 B4L1U3 A0A0P8Y7G7 B3N0C8 A0A1B0FNM8 A0A0L0C993 A0A0M9A038 A0A3B0J8E7 A0A1J1IXR0 Q29IB8 A0A1A9UHP7 B4M1J5 B4JL61 A0A2S2NS56 A0A0M5J642 A0A154PLS7 A0A1W4VHZ0 B4Q1W1 B3NW89 B4IEU0 A0A2J7PMQ8 B4R639 Q86LG1 A0A1B0GN28 A0A1B6MGD0 A0A158NBF0 A0A067RB77 A0A0C9RL33 A0A1S4EW92 A0A1A9WF90 A0A1D2MTK5 A0A232FIR5 B0WIZ9 K7IP12 A0A1D2N8C1 A0A226EXG1 A0A1I8N2I2 A0A1W4XR61 A0A1I8MQ88 A0A2W1BGB8 A0A139WCM4 B0WDC8 A0A226DYB2 A0A182GC76 B0WDC6 D6WXM7 A0A1D2MEU0 A0A226EYV0 A0A3S2NPU4 Q17NS9 A0A194PW83 A0A226F6M1 A0A1I8PEH0 A0A182MXN6 A0A1J1HMY9 A0A0L7L5Y6 A0A1Y1L465 Q17MX3 B0X0S1 A0A1I8PI06 N6U8E9 A0A1S4EX90 A0A182GRH0 T1H366 U4UGG3 Q9VS73 B4PC79 B3NFC4 A0A0J9RQW9 B4HV75 A0A182H7C8 A0A084WQC5 A0A1I8NHK4 B0WKI1 A0A1B0GL64 A0A226F2X9 A0A1S4G1A5 A0A226EYJ2 W5JCG6 B4J292 B3M5Z2 Q2M133 B4GUR0 A0A1B0GH50 A0A0L7LLW2 A0A0Q9WPD7 D6WXM6 A0A1I8N4A3 A0A182WEP1
Pubmed
24508170
30249741
25315136
17994087
26108605
15632085
+ More
17550304 12571352 10731132 12537572 12848938 20445050 20364123 21347285 24845553 17510324 27289101 28648823 20075255 28756777 18362917 19820115 26483478 26354079 26227816 28004739 23537049 12537568 12537573 12537574 16110336 17569856 17569867 22936249 24438588 20920257 23761445
17550304 12571352 10731132 12537572 12848938 20445050 20364123 21347285 24845553 17510324 27289101 28648823 20075255 28756777 18362917 19820115 26483478 26354079 26227816 28004739 23537049 12537568 12537573 12537574 16110336 17569856 17569867 22936249 24438588 20920257 23761445
EMBL
NWSH01000320
PCG77501.1
RSAL01000051
RVE50284.1
ODYU01000646
SOQ35776.1
+ More
KK107250 EZA54394.1 QOIP01000002 RLU25553.1 JXJN01002618 CH964239 EDW81987.1 CH933810 EDW06746.1 CH902640 KPU77367.1 EDV38332.1 CCAG010007674 JRES01000741 KNC28817.1 KQ435803 KOX73079.1 OUUW01000003 SPP78135.1 CVRI01000064 CRL05059.1 CH379063 EAL32735.2 CH940651 EDW65549.1 CH916370 EDW00314.1 GGMR01007365 MBY19984.1 CP012528 ALC49086.1 KQ434948 KZC12404.1 CM000162 EDX02536.1 CH954180 EDV46428.1 CH480832 EDW46194.1 NEVH01023960 PNF17623.1 CM000366 EDX18143.1 AY226552 AY226553 AE014298 BT122074 AAF48671.3 AAO47378.1 ADE06680.1 AJVK01027922 AJVK01027923 AJVK01027924 GEBQ01005015 JAT34962.1 ADTU01000293 KK852572 KDR21106.1 GBYB01008935 JAG78702.1 LJIJ01000549 ODM96336.1 NNAY01000169 OXU30358.1 DS231954 EDS28860.1 LJIJ01000149 ODN01494.1 LNIX01000001 OXA61306.1 KZ150082 PZC73838.1 KQ971362 KYB25718.1 DS231896 EDS44441.1 LNIX01000009 OXA50442.1 JXUM01054684 KQ561832 KXJ77400.1 EDS44439.1 EFA08874.1 LJIJ01001558 ODM91404.1 OXA61991.1 RSAL01000014 RVE53205.1 CH477196 EAT48395.1 KQ459591 KPI97019.1 OXA64506.1 CVRI01000004 CRK87601.1 JTDY01002789 KOB70726.1 GEZM01065387 JAV68489.1 CH477203 EAT48029.1 DS232244 EDS38313.1 APGK01038999 APGK01039000 KB740966 ENN76926.1 JXUM01082415 KQ563311 KXJ74085.1 CAQQ02171020 KB632186 ERL89696.1 AE014296 AAF50555.3 CM000159 EDW93764.1 CH954178 EDV50466.2 CM002912 KMY98102.1 CH480817 EDW50846.1 JXUM01116018 KQ565940 KXJ70514.1 ATLV01025482 KE525389 KFB52419.1 DS231972 EDS29857.1 AJWK01034404 AJWK01034405 OXA63838.1 OXA62672.1 ADMH02001572 ETN62032.1 CH916366 EDV97043.1 CH902618 EDV40708.1 CH379069 EAL30743.2 CH479191 EDW26343.1 AJWK01003969 AJWK01003970 AJWK01003971 AJWK01003972 JTDY01000591 KOB76548.1 KRF94123.1 EFA08873.2
KK107250 EZA54394.1 QOIP01000002 RLU25553.1 JXJN01002618 CH964239 EDW81987.1 CH933810 EDW06746.1 CH902640 KPU77367.1 EDV38332.1 CCAG010007674 JRES01000741 KNC28817.1 KQ435803 KOX73079.1 OUUW01000003 SPP78135.1 CVRI01000064 CRL05059.1 CH379063 EAL32735.2 CH940651 EDW65549.1 CH916370 EDW00314.1 GGMR01007365 MBY19984.1 CP012528 ALC49086.1 KQ434948 KZC12404.1 CM000162 EDX02536.1 CH954180 EDV46428.1 CH480832 EDW46194.1 NEVH01023960 PNF17623.1 CM000366 EDX18143.1 AY226552 AY226553 AE014298 BT122074 AAF48671.3 AAO47378.1 ADE06680.1 AJVK01027922 AJVK01027923 AJVK01027924 GEBQ01005015 JAT34962.1 ADTU01000293 KK852572 KDR21106.1 GBYB01008935 JAG78702.1 LJIJ01000549 ODM96336.1 NNAY01000169 OXU30358.1 DS231954 EDS28860.1 LJIJ01000149 ODN01494.1 LNIX01000001 OXA61306.1 KZ150082 PZC73838.1 KQ971362 KYB25718.1 DS231896 EDS44441.1 LNIX01000009 OXA50442.1 JXUM01054684 KQ561832 KXJ77400.1 EDS44439.1 EFA08874.1 LJIJ01001558 ODM91404.1 OXA61991.1 RSAL01000014 RVE53205.1 CH477196 EAT48395.1 KQ459591 KPI97019.1 OXA64506.1 CVRI01000004 CRK87601.1 JTDY01002789 KOB70726.1 GEZM01065387 JAV68489.1 CH477203 EAT48029.1 DS232244 EDS38313.1 APGK01038999 APGK01039000 KB740966 ENN76926.1 JXUM01082415 KQ563311 KXJ74085.1 CAQQ02171020 KB632186 ERL89696.1 AE014296 AAF50555.3 CM000159 EDW93764.1 CH954178 EDV50466.2 CM002912 KMY98102.1 CH480817 EDW50846.1 JXUM01116018 KQ565940 KXJ70514.1 ATLV01025482 KE525389 KFB52419.1 DS231972 EDS29857.1 AJWK01034404 AJWK01034405 OXA63838.1 OXA62672.1 ADMH02001572 ETN62032.1 CH916366 EDV97043.1 CH902618 EDV40708.1 CH379069 EAL30743.2 CH479191 EDW26343.1 AJWK01003969 AJWK01003970 AJWK01003971 AJWK01003972 JTDY01000591 KOB76548.1 KRF94123.1 EFA08873.2
Proteomes
UP000218220
UP000283053
UP000053097
UP000279307
UP000095300
UP000092460
+ More
UP000095301 UP000007798 UP000092443 UP000009192 UP000007801 UP000092444 UP000037069 UP000053105 UP000268350 UP000183832 UP000001819 UP000078200 UP000008792 UP000001070 UP000092553 UP000076502 UP000192221 UP000002282 UP000008711 UP000001292 UP000235965 UP000000304 UP000000803 UP000092462 UP000005205 UP000027135 UP000091820 UP000094527 UP000215335 UP000002320 UP000002358 UP000198287 UP000192223 UP000007266 UP000069940 UP000249989 UP000008820 UP000053268 UP000075884 UP000037510 UP000019118 UP000015102 UP000030742 UP000030765 UP000092461 UP000000673 UP000008744 UP000075920
UP000095301 UP000007798 UP000092443 UP000009192 UP000007801 UP000092444 UP000037069 UP000053105 UP000268350 UP000183832 UP000001819 UP000078200 UP000008792 UP000001070 UP000092553 UP000076502 UP000192221 UP000002282 UP000008711 UP000001292 UP000235965 UP000000304 UP000000803 UP000092462 UP000005205 UP000027135 UP000091820 UP000094527 UP000215335 UP000002320 UP000002358 UP000198287 UP000192223 UP000007266 UP000069940 UP000249989 UP000008820 UP000053268 UP000075884 UP000037510 UP000019118 UP000015102 UP000030742 UP000030765 UP000092461 UP000000673 UP000008744 UP000075920
Interpro
IPR001873
ENaC
+ More
IPR020903 ENaC_CS
IPR027417 P-loop_NTPase
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR014014 RNA_helicase_DEAD_Q_motif
IPR011545 DEAD/DEAH_box_helicase_dom
IPR042097 Aminopeptidase_N-like_N
IPR014782 Peptidase_M1_dom
IPR024571 ERAP1-like_C_dom
IPR033581 APN2
IPR001930 Peptidase_M1
IPR034016 M1_APN-typ
IPR020903 ENaC_CS
IPR027417 P-loop_NTPase
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR014014 RNA_helicase_DEAD_Q_motif
IPR011545 DEAD/DEAH_box_helicase_dom
IPR042097 Aminopeptidase_N-like_N
IPR014782 Peptidase_M1_dom
IPR024571 ERAP1-like_C_dom
IPR033581 APN2
IPR001930 Peptidase_M1
IPR034016 M1_APN-typ
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
A0A2A4JZR3
A0A437BIK4
A0A2H1V4S6
A0A026WH41
A0A3L8DZA0
A0A1I8NLF6
+ More
A0A1B0ARX1 A0A1I8MB58 A0A1I8M676 B4NE18 A0A1A9YCN5 B4L1U3 A0A0P8Y7G7 B3N0C8 A0A1B0FNM8 A0A0L0C993 A0A0M9A038 A0A3B0J8E7 A0A1J1IXR0 Q29IB8 A0A1A9UHP7 B4M1J5 B4JL61 A0A2S2NS56 A0A0M5J642 A0A154PLS7 A0A1W4VHZ0 B4Q1W1 B3NW89 B4IEU0 A0A2J7PMQ8 B4R639 Q86LG1 A0A1B0GN28 A0A1B6MGD0 A0A158NBF0 A0A067RB77 A0A0C9RL33 A0A1S4EW92 A0A1A9WF90 A0A1D2MTK5 A0A232FIR5 B0WIZ9 K7IP12 A0A1D2N8C1 A0A226EXG1 A0A1I8N2I2 A0A1W4XR61 A0A1I8MQ88 A0A2W1BGB8 A0A139WCM4 B0WDC8 A0A226DYB2 A0A182GC76 B0WDC6 D6WXM7 A0A1D2MEU0 A0A226EYV0 A0A3S2NPU4 Q17NS9 A0A194PW83 A0A226F6M1 A0A1I8PEH0 A0A182MXN6 A0A1J1HMY9 A0A0L7L5Y6 A0A1Y1L465 Q17MX3 B0X0S1 A0A1I8PI06 N6U8E9 A0A1S4EX90 A0A182GRH0 T1H366 U4UGG3 Q9VS73 B4PC79 B3NFC4 A0A0J9RQW9 B4HV75 A0A182H7C8 A0A084WQC5 A0A1I8NHK4 B0WKI1 A0A1B0GL64 A0A226F2X9 A0A1S4G1A5 A0A226EYJ2 W5JCG6 B4J292 B3M5Z2 Q2M133 B4GUR0 A0A1B0GH50 A0A0L7LLW2 A0A0Q9WPD7 D6WXM6 A0A1I8N4A3 A0A182WEP1
A0A1B0ARX1 A0A1I8MB58 A0A1I8M676 B4NE18 A0A1A9YCN5 B4L1U3 A0A0P8Y7G7 B3N0C8 A0A1B0FNM8 A0A0L0C993 A0A0M9A038 A0A3B0J8E7 A0A1J1IXR0 Q29IB8 A0A1A9UHP7 B4M1J5 B4JL61 A0A2S2NS56 A0A0M5J642 A0A154PLS7 A0A1W4VHZ0 B4Q1W1 B3NW89 B4IEU0 A0A2J7PMQ8 B4R639 Q86LG1 A0A1B0GN28 A0A1B6MGD0 A0A158NBF0 A0A067RB77 A0A0C9RL33 A0A1S4EW92 A0A1A9WF90 A0A1D2MTK5 A0A232FIR5 B0WIZ9 K7IP12 A0A1D2N8C1 A0A226EXG1 A0A1I8N2I2 A0A1W4XR61 A0A1I8MQ88 A0A2W1BGB8 A0A139WCM4 B0WDC8 A0A226DYB2 A0A182GC76 B0WDC6 D6WXM7 A0A1D2MEU0 A0A226EYV0 A0A3S2NPU4 Q17NS9 A0A194PW83 A0A226F6M1 A0A1I8PEH0 A0A182MXN6 A0A1J1HMY9 A0A0L7L5Y6 A0A1Y1L465 Q17MX3 B0X0S1 A0A1I8PI06 N6U8E9 A0A1S4EX90 A0A182GRH0 T1H366 U4UGG3 Q9VS73 B4PC79 B3NFC4 A0A0J9RQW9 B4HV75 A0A182H7C8 A0A084WQC5 A0A1I8NHK4 B0WKI1 A0A1B0GL64 A0A226F2X9 A0A1S4G1A5 A0A226EYJ2 W5JCG6 B4J292 B3M5Z2 Q2M133 B4GUR0 A0A1B0GH50 A0A0L7LLW2 A0A0Q9WPD7 D6WXM6 A0A1I8N4A3 A0A182WEP1
Ontologies
GO
PANTHER
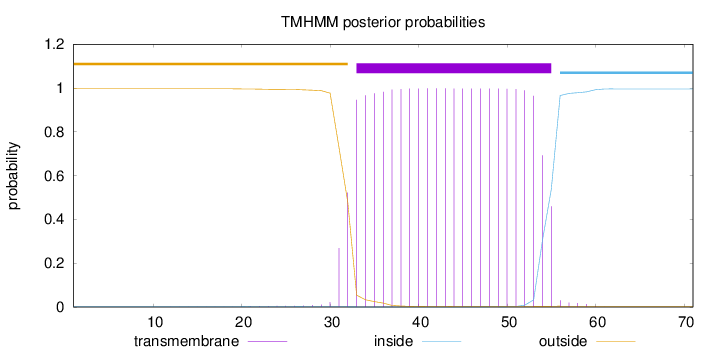
Topology
Subcellular location
Cell membrane
Length:
71
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.88583
Exp number, first 60 AAs:
22.88519
Total prob of N-in:
0.00340
POSSIBLE N-term signal
sequence
outside
1 - 32
TMhelix
33 - 55
inside
56 - 71
Population Genetic Test Statistics
Pi
126.794455
Theta
121.982349
Tajima's D
-0.339952
CLR
0.053702
CSRT
0.28053597320134
Interpretation
Uncertain
