Gene
KWMTBOMO09615
Pre Gene Modal
BGIBMGA012849
Annotation
PREDICTED:_sodium_channel_protein_Nach-like_isoform_X1_[Amyelois_transitella]
Full name
Pickpocket protein 28
Location in the cell
PlasmaMembrane Reliability : 2.24
Sequence
CDS
ATGACCAAAATTCAACAAGCCCCACAATCAAAGAACCAAAGTTTAGATGACGAAGAACCTCTGTCTTTTGCGAGTGACATAAGGCAATATCTACAATGTTCCACGTTGCACGGTCTTCGGTATATTGCCGAGAAGAAACTGACTTGGTTCGAAAGGTTCTTCTGGCTGGCAGCTTTTAGCTCCTCCTTAATTTGCGCTGGATTCTTTATTCTAAACGTGTATTCGAAATGGCGGATGTCGCCAATGATAGTGAGCATCAATCCTGAAAATATGCCTTTGCATGGCTTGCCTTTCCCAGCTATAACCATTTGCAATGTTAATCAAGCAAAGAAATCTATCGCCGAAGGATACCAAAAATACGGGTGA
Protein
MTKIQQAPQSKNQSLDDEEPLSFASDIRQYLQCSTLHGLRYIAEKKLTWFERFFWLAAFSSSLICAGFFILNVYSKWRMSPMIVSINPENMPLHGLPFPAITICNVNQAKKSIAEGYQKYG
Summary
Description
Osmosensitive ion channel that mediates the cellular and behavioral response to water. Plays an essential role in gustatory water reception. Part of a complex that plays a role in tracheal liquid clearance. Probable role in sodium transport.
Similarity
Belongs to the amiloride-sensitive sodium channel (TC 1.A.6) family.
Belongs to the two pore domain potassium channel (TC 1.A.1.8) family.
Belongs to the two pore domain potassium channel (TC 1.A.1.8) family.
Keywords
Cell membrane
Complete proteome
Ion channel
Ion transport
Membrane
Reference proteome
Sodium
Sodium channel
Sodium transport
Transmembrane
Transmembrane helix
Transport
Feature
chain Pickpocket protein 28
Uniprot
A0A2H1V4S6
A0A437BIK4
A0A2A4JZR3
A0A212FML8
A0A0N1PJJ9
A0A0N1PJY5
+ More
N6T2I8 U4U735 E2BNL2 A0A226DRB1 B3N2P2 A0A182YAL4 A0A0M5J642 E9IWN6 F4X372 W5J1E9 A0A2S2R4W4 A0A182GC76 A0A151WEN4 B3NW89 A0A0L7QMQ9 A0A195BDN0 A0A067RB77 A0A1I8NLF6 B4JL61 J9LZ57 A0A1B0ARX1 A0A158NBF0 B4IEU0 B4R639 A0A2A3EG41 A0A182IM43 A0A154PLS7 A0A0M9A038 B3N0C8 A0A0P8Y7G7 A0A1W4XBD2 E2A269 B4P7Y1 B4QH60 A0A182WSG5 A0A1A9YCN5 A0A088A4H4 A0A182Q3R4 A0A0J9RIX3 A0A182VK33 B3NNP7 B4I849 A0A0P6A4R8 A0A1B0ABX7 A0A0P5ZYH3 Q7PKT6 A0A182HLB5 Q8T5H4 A0A1B0FNM8 A0A182KFD0 A0A195FFW2 A0A3B0J8E7 D7EL39 A0A2P8XRE9 A0A232FIR5 A0A1A9WF90 A0A0L0C993 A0A182L9P3 Q8MU73 A0A1A9UHP7 A0A182UKS5 A0A1W4VP24 Q9W250 A0A182FA76 B4L1U3 A0A226DRR1 A0A0P6CR49 A0A182VZA7 A0A3Q0J677 A0A1W4VHZ0 A0A164V2P5 A0A182R635 A0A0L0CLL3 A0A182PGQ2 A0A0P6HZM3 Q17NS9 A0A182NS56 K7IP12 A0A1S4EW92 Q86LG1 A0A182TA96 A0A182MTE6 A0A026WH41 A0A1B0GH45 A0A3L8DZA0 A0A2P8Z373 A0A1J1IXR0 A0A0A9XE52 A0A1I8MUU6 B4M1J5 B4Q1W1
N6T2I8 U4U735 E2BNL2 A0A226DRB1 B3N2P2 A0A182YAL4 A0A0M5J642 E9IWN6 F4X372 W5J1E9 A0A2S2R4W4 A0A182GC76 A0A151WEN4 B3NW89 A0A0L7QMQ9 A0A195BDN0 A0A067RB77 A0A1I8NLF6 B4JL61 J9LZ57 A0A1B0ARX1 A0A158NBF0 B4IEU0 B4R639 A0A2A3EG41 A0A182IM43 A0A154PLS7 A0A0M9A038 B3N0C8 A0A0P8Y7G7 A0A1W4XBD2 E2A269 B4P7Y1 B4QH60 A0A182WSG5 A0A1A9YCN5 A0A088A4H4 A0A182Q3R4 A0A0J9RIX3 A0A182VK33 B3NNP7 B4I849 A0A0P6A4R8 A0A1B0ABX7 A0A0P5ZYH3 Q7PKT6 A0A182HLB5 Q8T5H4 A0A1B0FNM8 A0A182KFD0 A0A195FFW2 A0A3B0J8E7 D7EL39 A0A2P8XRE9 A0A232FIR5 A0A1A9WF90 A0A0L0C993 A0A182L9P3 Q8MU73 A0A1A9UHP7 A0A182UKS5 A0A1W4VP24 Q9W250 A0A182FA76 B4L1U3 A0A226DRR1 A0A0P6CR49 A0A182VZA7 A0A3Q0J677 A0A1W4VHZ0 A0A164V2P5 A0A182R635 A0A0L0CLL3 A0A182PGQ2 A0A0P6HZM3 Q17NS9 A0A182NS56 K7IP12 A0A1S4EW92 Q86LG1 A0A182TA96 A0A182MTE6 A0A026WH41 A0A1B0GH45 A0A3L8DZA0 A0A2P8Z373 A0A1J1IXR0 A0A0A9XE52 A0A1I8MUU6 B4M1J5 B4Q1W1
Pubmed
22118469
26354079
23537049
20798317
17994087
25244985
+ More
21282665 21719571 20920257 23761445 26483478 24845553 21347285 17550304 22936249 12364791 14747013 17210077 12060762 18362917 19820115 29403074 28648823 26108605 20966253 10731132 12537568 12537572 12537573 12537574 12571352 16110336 17569856 17569867 26109357 26109356 17510324 20075255 12848938 20445050 20364123 24508170 30249741 25401762 25315136
21282665 21719571 20920257 23761445 26483478 24845553 21347285 17550304 22936249 12364791 14747013 17210077 12060762 18362917 19820115 29403074 28648823 26108605 20966253 10731132 12537568 12537572 12537573 12537574 12571352 16110336 17569856 17569867 26109357 26109356 17510324 20075255 12848938 20445050 20364123 24508170 30249741 25401762 25315136
EMBL
ODYU01000646
SOQ35776.1
RSAL01000051
RVE50284.1
NWSH01000320
PCG77501.1
+ More
AGBW02007651 OWR54981.1 LADI01006439 KPJ20638.1 KQ460208 KPJ16605.1 APGK01054977 KB741256 ENN71763.1 KB631749 ERL85770.1 GL449414 EFN82791.1 LNIX01000013 OXA47207.1 CH905151 EDV29966.2 CP012528 ALC49086.1 GL766597 EFZ14977.1 GL888609 EGI59053.1 ADMH02002183 ETN57987.1 GGMS01015801 MBY85004.1 JXUM01054684 KQ561832 KXJ77400.1 KQ983238 KYQ46255.1 CH954180 EDV46428.1 KQ414885 KOC59880.1 KQ976509 KYM82676.1 KK852572 KDR21106.1 CH916370 EDW00314.1 ABLF02032360 ABLF02032364 JXJN01002618 ADTU01000293 CH480832 EDW46194.1 CM000366 EDX18143.1 KZ288266 PBC30262.1 KQ434948 KZC12404.1 KQ435803 KOX73079.1 CH902640 EDV38332.1 KPU77367.1 GL435922 EFN72474.1 CM000158 EDW92136.1 CM000362 EDX08206.1 AXCN02000266 CM002911 KMY95792.1 CH954179 EDV55604.1 CH480824 EDW56774.1 GDIP01039413 JAM64302.1 GDIP01037965 JAM65750.1 AAAB01008987 EAA43142.2 APCN01000890 AJ439398 CAD28127.1 CCAG010007674 KQ981606 KYN39585.1 OUUW01000003 SPP78135.1 KQ973192 EFA11834.2 PYGN01001481 PSN34583.1 NNAY01000169 OXU30358.1 JRES01000741 KNC28817.1 AJ441131 CAD29634.1 AE013599 AY226543 AAF46849.2 AAO47369.1 CH933810 EDW06746.1 OXA47357.1 GDIP01004478 JAM99237.1 LRGB01001463 KZS11907.1 JRES01000319 KNC32334.1 GDIQ01011901 JAN82836.1 CH477196 EAT48395.1 AY226552 AY226553 AE014298 BT122074 AAF48671.3 AAO47378.1 ADE06680.1 AXCM01013744 AXCM01013745 KK107250 EZA54394.1 AJWK01003962 AJWK01003963 AJWK01003964 QOIP01000002 RLU25553.1 PYGN01000218 PSN50939.1 CVRI01000064 CRL05059.1 GBHO01025335 JAG18269.1 CH940651 EDW65549.1 CM000162 EDX02536.1
AGBW02007651 OWR54981.1 LADI01006439 KPJ20638.1 KQ460208 KPJ16605.1 APGK01054977 KB741256 ENN71763.1 KB631749 ERL85770.1 GL449414 EFN82791.1 LNIX01000013 OXA47207.1 CH905151 EDV29966.2 CP012528 ALC49086.1 GL766597 EFZ14977.1 GL888609 EGI59053.1 ADMH02002183 ETN57987.1 GGMS01015801 MBY85004.1 JXUM01054684 KQ561832 KXJ77400.1 KQ983238 KYQ46255.1 CH954180 EDV46428.1 KQ414885 KOC59880.1 KQ976509 KYM82676.1 KK852572 KDR21106.1 CH916370 EDW00314.1 ABLF02032360 ABLF02032364 JXJN01002618 ADTU01000293 CH480832 EDW46194.1 CM000366 EDX18143.1 KZ288266 PBC30262.1 KQ434948 KZC12404.1 KQ435803 KOX73079.1 CH902640 EDV38332.1 KPU77367.1 GL435922 EFN72474.1 CM000158 EDW92136.1 CM000362 EDX08206.1 AXCN02000266 CM002911 KMY95792.1 CH954179 EDV55604.1 CH480824 EDW56774.1 GDIP01039413 JAM64302.1 GDIP01037965 JAM65750.1 AAAB01008987 EAA43142.2 APCN01000890 AJ439398 CAD28127.1 CCAG010007674 KQ981606 KYN39585.1 OUUW01000003 SPP78135.1 KQ973192 EFA11834.2 PYGN01001481 PSN34583.1 NNAY01000169 OXU30358.1 JRES01000741 KNC28817.1 AJ441131 CAD29634.1 AE013599 AY226543 AAF46849.2 AAO47369.1 CH933810 EDW06746.1 OXA47357.1 GDIP01004478 JAM99237.1 LRGB01001463 KZS11907.1 JRES01000319 KNC32334.1 GDIQ01011901 JAN82836.1 CH477196 EAT48395.1 AY226552 AY226553 AE014298 BT122074 AAF48671.3 AAO47378.1 ADE06680.1 AXCM01013744 AXCM01013745 KK107250 EZA54394.1 AJWK01003962 AJWK01003963 AJWK01003964 QOIP01000002 RLU25553.1 PYGN01000218 PSN50939.1 CVRI01000064 CRL05059.1 GBHO01025335 JAG18269.1 CH940651 EDW65549.1 CM000162 EDX02536.1
Proteomes
UP000283053
UP000218220
UP000007151
UP000053268
UP000053240
UP000019118
+ More
UP000030742 UP000008237 UP000198287 UP000007801 UP000076408 UP000092553 UP000007755 UP000000673 UP000069940 UP000249989 UP000075809 UP000008711 UP000053825 UP000078540 UP000027135 UP000095300 UP000001070 UP000007819 UP000092460 UP000005205 UP000001292 UP000000304 UP000242457 UP000075880 UP000076502 UP000053105 UP000192223 UP000000311 UP000002282 UP000076407 UP000092443 UP000005203 UP000075886 UP000075903 UP000092445 UP000007062 UP000075840 UP000092444 UP000075881 UP000078541 UP000268350 UP000007266 UP000245037 UP000215335 UP000091820 UP000037069 UP000075882 UP000078200 UP000075902 UP000192221 UP000000803 UP000069272 UP000009192 UP000075920 UP000079169 UP000076858 UP000075900 UP000075885 UP000008820 UP000075884 UP000002358 UP000075901 UP000075883 UP000053097 UP000092461 UP000279307 UP000183832 UP000095301 UP000008792
UP000030742 UP000008237 UP000198287 UP000007801 UP000076408 UP000092553 UP000007755 UP000000673 UP000069940 UP000249989 UP000075809 UP000008711 UP000053825 UP000078540 UP000027135 UP000095300 UP000001070 UP000007819 UP000092460 UP000005205 UP000001292 UP000000304 UP000242457 UP000075880 UP000076502 UP000053105 UP000192223 UP000000311 UP000002282 UP000076407 UP000092443 UP000005203 UP000075886 UP000075903 UP000092445 UP000007062 UP000075840 UP000092444 UP000075881 UP000078541 UP000268350 UP000007266 UP000245037 UP000215335 UP000091820 UP000037069 UP000075882 UP000078200 UP000075902 UP000192221 UP000000803 UP000069272 UP000009192 UP000075920 UP000079169 UP000076858 UP000075900 UP000075885 UP000008820 UP000075884 UP000002358 UP000075901 UP000075883 UP000053097 UP000092461 UP000279307 UP000183832 UP000095301 UP000008792
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
A0A2H1V4S6
A0A437BIK4
A0A2A4JZR3
A0A212FML8
A0A0N1PJJ9
A0A0N1PJY5
+ More
N6T2I8 U4U735 E2BNL2 A0A226DRB1 B3N2P2 A0A182YAL4 A0A0M5J642 E9IWN6 F4X372 W5J1E9 A0A2S2R4W4 A0A182GC76 A0A151WEN4 B3NW89 A0A0L7QMQ9 A0A195BDN0 A0A067RB77 A0A1I8NLF6 B4JL61 J9LZ57 A0A1B0ARX1 A0A158NBF0 B4IEU0 B4R639 A0A2A3EG41 A0A182IM43 A0A154PLS7 A0A0M9A038 B3N0C8 A0A0P8Y7G7 A0A1W4XBD2 E2A269 B4P7Y1 B4QH60 A0A182WSG5 A0A1A9YCN5 A0A088A4H4 A0A182Q3R4 A0A0J9RIX3 A0A182VK33 B3NNP7 B4I849 A0A0P6A4R8 A0A1B0ABX7 A0A0P5ZYH3 Q7PKT6 A0A182HLB5 Q8T5H4 A0A1B0FNM8 A0A182KFD0 A0A195FFW2 A0A3B0J8E7 D7EL39 A0A2P8XRE9 A0A232FIR5 A0A1A9WF90 A0A0L0C993 A0A182L9P3 Q8MU73 A0A1A9UHP7 A0A182UKS5 A0A1W4VP24 Q9W250 A0A182FA76 B4L1U3 A0A226DRR1 A0A0P6CR49 A0A182VZA7 A0A3Q0J677 A0A1W4VHZ0 A0A164V2P5 A0A182R635 A0A0L0CLL3 A0A182PGQ2 A0A0P6HZM3 Q17NS9 A0A182NS56 K7IP12 A0A1S4EW92 Q86LG1 A0A182TA96 A0A182MTE6 A0A026WH41 A0A1B0GH45 A0A3L8DZA0 A0A2P8Z373 A0A1J1IXR0 A0A0A9XE52 A0A1I8MUU6 B4M1J5 B4Q1W1
N6T2I8 U4U735 E2BNL2 A0A226DRB1 B3N2P2 A0A182YAL4 A0A0M5J642 E9IWN6 F4X372 W5J1E9 A0A2S2R4W4 A0A182GC76 A0A151WEN4 B3NW89 A0A0L7QMQ9 A0A195BDN0 A0A067RB77 A0A1I8NLF6 B4JL61 J9LZ57 A0A1B0ARX1 A0A158NBF0 B4IEU0 B4R639 A0A2A3EG41 A0A182IM43 A0A154PLS7 A0A0M9A038 B3N0C8 A0A0P8Y7G7 A0A1W4XBD2 E2A269 B4P7Y1 B4QH60 A0A182WSG5 A0A1A9YCN5 A0A088A4H4 A0A182Q3R4 A0A0J9RIX3 A0A182VK33 B3NNP7 B4I849 A0A0P6A4R8 A0A1B0ABX7 A0A0P5ZYH3 Q7PKT6 A0A182HLB5 Q8T5H4 A0A1B0FNM8 A0A182KFD0 A0A195FFW2 A0A3B0J8E7 D7EL39 A0A2P8XRE9 A0A232FIR5 A0A1A9WF90 A0A0L0C993 A0A182L9P3 Q8MU73 A0A1A9UHP7 A0A182UKS5 A0A1W4VP24 Q9W250 A0A182FA76 B4L1U3 A0A226DRR1 A0A0P6CR49 A0A182VZA7 A0A3Q0J677 A0A1W4VHZ0 A0A164V2P5 A0A182R635 A0A0L0CLL3 A0A182PGQ2 A0A0P6HZM3 Q17NS9 A0A182NS56 K7IP12 A0A1S4EW92 Q86LG1 A0A182TA96 A0A182MTE6 A0A026WH41 A0A1B0GH45 A0A3L8DZA0 A0A2P8Z373 A0A1J1IXR0 A0A0A9XE52 A0A1I8MUU6 B4M1J5 B4Q1W1
PDB
6AVE
E-value=0.0001208,
Score=101
Ontologies
GO
PANTHER
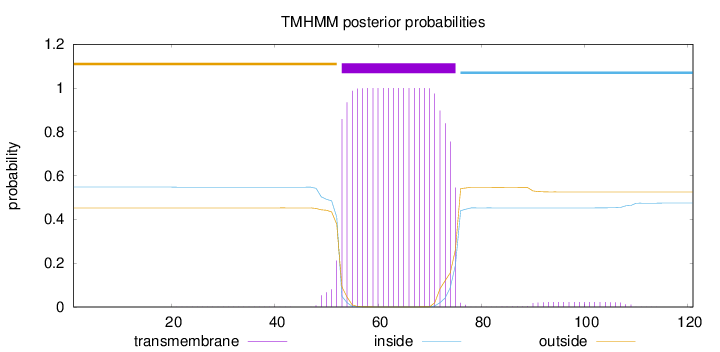
Topology
Subcellular location
Cell membrane
Length:
121
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.66604
Exp number, first 60 AAs:
8.20068
Total prob of N-in:
0.54752
outside
1 - 52
TMhelix
53 - 75
inside
76 - 121
Population Genetic Test Statistics
Pi
235.68783
Theta
201.70917
Tajima's D
0.510794
CLR
0.047479
CSRT
0.516174191290436
Interpretation
Uncertain
