Gene
KWMTBOMO09607
Annotation
PREDICTED:_uncharacterized_protein_LOC105842033_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.477
Sequence
CDS
ATGAAAAACAGGACAATCAAGATGGACAAGCCACGAAGGAAATATAAAGATTACGACGACAGCTTGCTAGAAATTGCGGTAGGACTCGTAGAAAATAAGAATATTTCGTCATACGAGGCAGAAAAACAATTTGGTATCCCTCGCCGCACGATTTTAAATAAGGTCAAACAAAGACACACAAAAAGAGTTGGCTGTCCTACCAAACTGCCACTAGCTGATGAAAAGAAAATTACAAGCTGTTTGATTCTATGTGGAGAATATGGGTATCCCCTTACCTGCTTTGAACTAAGGTCAATAGTACACGACTATTTAGTTAAAAATGGACTCACTTATCTTTTTAATAACAAAATGCCCGGCCAGAAGTGGGTAAGGAACTTTTTGAAACGCAACAAAAATATTCTGACTGTGCGGTCTACAACAAATATAAAGGAATCTCGGGCCAGAAAAAGCATACAAGACTACAATACTTATTTTGAGAACCTTAAAATTTCTCTTCATAATGTTAAACCAGATGATATAGTGAACTATGACGAGACAAATTTTTCTGACAACCCGGGATCCCAAAAATGCATATTTAAAGCAGGCACTAAACATCCAAATAAAACTTTAAATAGCACCAAAACTGCAATATCAGTCATGTTTTCGGCTACAGCCAGTGGAGTAATATTCCCACCGTATGTGGTATATAAAGCTGATAATTTATGGACAAACTGGTGTACTAATGGTCCTAAAGGAACCAGATATAACAAAACCAAATCTGGCTGGTTTGATTCGGAAACGTTCGAAGATTGGTTTAATACCATAATTATTCCATGGGCAAAGAAATCTACAGATCCTAAGGTCTTGATCGGAGACAATTTATCATCACATATAAATATAAGCATAATAACAAAGTGTCAAGAAAATAATATAAGATTTATTTTCTTGCCACCCAATTCGACACATCATACTCAACCTCTCGATGTGGCCTTTTTCGGCCCATTAAAGAGAGAATGGAGAAGAATTTTAATGGACTACAAAGTTCGACACCCAAGCGCAAATACCTTAAATAAACAAATATTCCCCGAATTACTAAATATACTACTGGATGCCTCGAAACTTACACAAAGTAAAAATATAATTAGCGGATTTCGTGCCACTGGGATACATCCTATTAATCCAGAAGCTGTACTAAAGAAATTGCCAGATTATCGCAGGCTTCAAACCGTGACTTATGACGAGACCTTTGTCAACTATTTACAAAATTTGAGAACTCCGGCTACCCATCAAAAAAGAACTGCAAATAAAAAACTAAATACAGAACCTGGAAAATCTGTAAGTGTTAATGATTTGAATGAAAAACTTAACAAACGGGCTAATAAAGTAAAAAATAATAAAAAGACAAGTACCATTACCGAAAAGATTCAGTTACAACCGGATATCGCAGACATTTCTACTGCTATTTTAGACCGTAATTTAAACAATATTTTATCAAATGCTAGACAAGATTTATCACCTCATTCAGATGATAGCAAACTTATTACTAATGTAAAAGTTCATGCTGAAGTACATAGAAATAAGGGTATGGATTCATTTGATATGAGCTCAGCTCTTGTCGATGGAGCAACAAGTGATTAA
Protein
MKNRTIKMDKPRRKYKDYDDSLLEIAVGLVENKNISSYEAEKQFGIPRRTILNKVKQRHTKRVGCPTKLPLADEKKITSCLILCGEYGYPLTCFELRSIVHDYLVKNGLTYLFNNKMPGQKWVRNFLKRNKNILTVRSTTNIKESRARKSIQDYNTYFENLKISLHNVKPDDIVNYDETNFSDNPGSQKCIFKAGTKHPNKTLNSTKTAISVMFSATASGVIFPPYVVYKADNLWTNWCTNGPKGTRYNKTKSGWFDSETFEDWFNTIIIPWAKKSTDPKVLIGDNLSSHINISIITKCQENNIRFIFLPPNSTHHTQPLDVAFFGPLKREWRRILMDYKVRHPSANTLNKQIFPELLNILLDASKLTQSKNIISGFRATGIHPINPEAVLKKLPDYRRLQTVTYDETFVNYLQNLRTPATHQKRTANKKLNTEPGKSVSVNDLNEKLNKRANKVKNNKKTSTITEKIQLQPDIADISTAILDRNLNNILSNARQDLSPHSDDSKLITNVKVHAEVHRNKGMDSFDMSSALVDGATSD
Summary
Uniprot
A0A3S2TIP6
A0A1E1WU51
A0A2W1BUJ3
A0A2A4IT23
A0A1Y1KZ51
X1XEK3
+ More
A0A1B6KLJ7 J9KC80 J9JX79 X1WLE6 J9JNH9 A0A1B6L0P8 A0A336M092 A0A1S4EG94 A0A3Q0J1E8 J9K6M6 X1WST4 A0A1Y1K537 J9LW80 A0A2S2R564 J9LWC9 A0A1B6GCB9 J9KAI8 H9JBQ2 J9M3V3 A0A2H8TJC7 J9K4L7 J9JPC0 A0A2S2NMJ8 A0A2P8YWM9 A0A2P8Z814 A0A1B6CLN0 B0XDM0 J9K1E0 J9M6Q1 J9KH76 J9K5J2 J9KE52 J9L3Y1 X1XT95 J9JJQ5 A0A1W7R625 A0A0J7KF01 J9LSN8 A0A1B6JFL2 A0A2W1BR16 K7JIQ4 A0A2A4JKM4 A0A2H8TYF0
A0A1B6KLJ7 J9KC80 J9JX79 X1WLE6 J9JNH9 A0A1B6L0P8 A0A336M092 A0A1S4EG94 A0A3Q0J1E8 J9K6M6 X1WST4 A0A1Y1K537 J9LW80 A0A2S2R564 J9LWC9 A0A1B6GCB9 J9KAI8 H9JBQ2 J9M3V3 A0A2H8TJC7 J9K4L7 J9JPC0 A0A2S2NMJ8 A0A2P8YWM9 A0A2P8Z814 A0A1B6CLN0 B0XDM0 J9K1E0 J9M6Q1 J9KH76 J9K5J2 J9KE52 J9L3Y1 X1XT95 J9JJQ5 A0A1W7R625 A0A0J7KF01 J9LSN8 A0A1B6JFL2 A0A2W1BR16 K7JIQ4 A0A2A4JKM4 A0A2H8TYF0
EMBL
RSAL01000111
RVE47096.1
GDQN01000733
JAT90321.1
KZ149957
PZC76450.1
+ More
NWSH01008222 PCG62646.1 GEZM01069098 JAV66644.1 ABLF02014120 ABLF02031335 GEBQ01027923 JAT12054.1 ABLF02031950 ABLF02024787 ABLF02042417 ABLF02024890 GEBQ01022752 JAT17225.1 UFQS01000373 UFQT01000373 SSX03288.1 SSX23654.1 ABLF02012638 ABLF02006951 GEZM01096654 GEZM01096605 JAV54775.1 ABLF02032408 ABLF02032409 ABLF02032413 GGMS01015966 MBY85169.1 ABLF02036293 ABLF02036295 ABLF02036300 ABLF02036303 ABLF02036307 GECZ01009826 JAS59943.1 ABLF02020969 ABLF02020971 BABH01024482 ABLF02005267 ABLF02005269 GFXV01002448 MBW14253.1 ABLF02035413 ABLF02021095 ABLF02021103 ABLF02021105 GGMR01005791 MBY18410.1 PYGN01000316 PSN48625.1 PYGN01000900 PYGN01000153 PSN39546.1 PSN52644.1 GEDC01023010 JAS14288.1 DS232767 EDS45530.1 ABLF02018386 ABLF02004276 ABLF02004277 ABLF02019754 ABLF02043119 ABLF02012639 ABLF02002783 ABLF02010048 ABLF02034252 ABLF02013132 ABLF02013134 ABLF02013135 ABLF02005217 GEHC01001021 JAV46624.1 LBMM01008491 KMQ88832.1 ABLF02009566 GECU01010063 JAS97643.1 KZ149928 PZC77478.1 AAZX01014011 NWSH01001259 PCG71952.1 GFXV01007016 MBW18821.1
NWSH01008222 PCG62646.1 GEZM01069098 JAV66644.1 ABLF02014120 ABLF02031335 GEBQ01027923 JAT12054.1 ABLF02031950 ABLF02024787 ABLF02042417 ABLF02024890 GEBQ01022752 JAT17225.1 UFQS01000373 UFQT01000373 SSX03288.1 SSX23654.1 ABLF02012638 ABLF02006951 GEZM01096654 GEZM01096605 JAV54775.1 ABLF02032408 ABLF02032409 ABLF02032413 GGMS01015966 MBY85169.1 ABLF02036293 ABLF02036295 ABLF02036300 ABLF02036303 ABLF02036307 GECZ01009826 JAS59943.1 ABLF02020969 ABLF02020971 BABH01024482 ABLF02005267 ABLF02005269 GFXV01002448 MBW14253.1 ABLF02035413 ABLF02021095 ABLF02021103 ABLF02021105 GGMR01005791 MBY18410.1 PYGN01000316 PSN48625.1 PYGN01000900 PYGN01000153 PSN39546.1 PSN52644.1 GEDC01023010 JAS14288.1 DS232767 EDS45530.1 ABLF02018386 ABLF02004276 ABLF02004277 ABLF02019754 ABLF02043119 ABLF02012639 ABLF02002783 ABLF02010048 ABLF02034252 ABLF02013132 ABLF02013134 ABLF02013135 ABLF02005217 GEHC01001021 JAV46624.1 LBMM01008491 KMQ88832.1 ABLF02009566 GECU01010063 JAS97643.1 KZ149928 PZC77478.1 AAZX01014011 NWSH01001259 PCG71952.1 GFXV01007016 MBW18821.1
Proteomes
PRIDE
Pfam
Interpro
IPR004875
DDE_SF_endonuclease_dom
+ More
IPR009057 Homeobox-like_sf
IPR007889 HTH_Psq
IPR008906 HATC_C_dom
IPR012337 RNaseH-like_sf
IPR006579 Pre_C2HC_dom
IPR007021 DUF659
IPR029526 PGBD
IPR027124 Swc5/CFDP2
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR011335 Restrct_endonuc-II-like
IPR011604 Exonuc_phg/RecB_C
IPR008042 Retrotrans_Pao
IPR006600 HTH_CenpB_DNA-bd_dom
IPR009057 Homeobox-like_sf
IPR007889 HTH_Psq
IPR008906 HATC_C_dom
IPR012337 RNaseH-like_sf
IPR006579 Pre_C2HC_dom
IPR007021 DUF659
IPR029526 PGBD
IPR027124 Swc5/CFDP2
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR011335 Restrct_endonuc-II-like
IPR011604 Exonuc_phg/RecB_C
IPR008042 Retrotrans_Pao
IPR006600 HTH_CenpB_DNA-bd_dom
Gene 3D
ProteinModelPortal
A0A3S2TIP6
A0A1E1WU51
A0A2W1BUJ3
A0A2A4IT23
A0A1Y1KZ51
X1XEK3
+ More
A0A1B6KLJ7 J9KC80 J9JX79 X1WLE6 J9JNH9 A0A1B6L0P8 A0A336M092 A0A1S4EG94 A0A3Q0J1E8 J9K6M6 X1WST4 A0A1Y1K537 J9LW80 A0A2S2R564 J9LWC9 A0A1B6GCB9 J9KAI8 H9JBQ2 J9M3V3 A0A2H8TJC7 J9K4L7 J9JPC0 A0A2S2NMJ8 A0A2P8YWM9 A0A2P8Z814 A0A1B6CLN0 B0XDM0 J9K1E0 J9M6Q1 J9KH76 J9K5J2 J9KE52 J9L3Y1 X1XT95 J9JJQ5 A0A1W7R625 A0A0J7KF01 J9LSN8 A0A1B6JFL2 A0A2W1BR16 K7JIQ4 A0A2A4JKM4 A0A2H8TYF0
A0A1B6KLJ7 J9KC80 J9JX79 X1WLE6 J9JNH9 A0A1B6L0P8 A0A336M092 A0A1S4EG94 A0A3Q0J1E8 J9K6M6 X1WST4 A0A1Y1K537 J9LW80 A0A2S2R564 J9LWC9 A0A1B6GCB9 J9KAI8 H9JBQ2 J9M3V3 A0A2H8TJC7 J9K4L7 J9JPC0 A0A2S2NMJ8 A0A2P8YWM9 A0A2P8Z814 A0A1B6CLN0 B0XDM0 J9K1E0 J9M6Q1 J9KH76 J9K5J2 J9KE52 J9L3Y1 X1XT95 J9JJQ5 A0A1W7R625 A0A0J7KF01 J9LSN8 A0A1B6JFL2 A0A2W1BR16 K7JIQ4 A0A2A4JKM4 A0A2H8TYF0
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
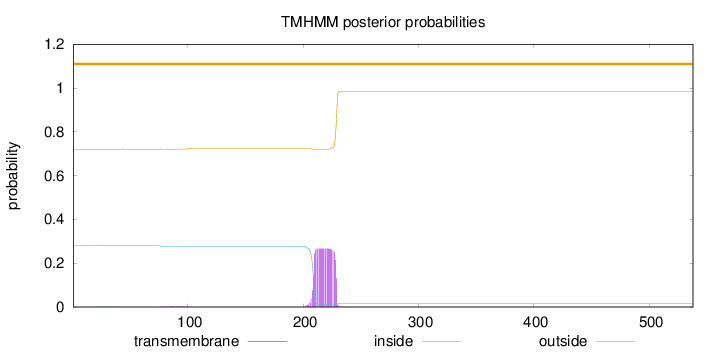
Length:
538
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.45731999999999
Exp number, first 60 AAs:
0.00657
Total prob of N-in:
0.27956
outside
1 - 538
Population Genetic Test Statistics
Pi
153.168609
Theta
170.431399
Tajima's D
-1.006767
CLR
175.230413
CSRT
0.136793160341983
Interpretation
Uncertain
