Gene
KWMTBOMO09606
Pre Gene Modal
BGIBMGA012843
Annotation
PREDICTED:_jmjC_domain-containing_histone_demethylation_protein_1_isoform_X3_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.209
Sequence
CDS
ATGTCGGAAACACTGCCGAATAATAAAAAACAAGTACCAAGACAGCTGCGTGAACGCAAGCACAGGAAGCTTTACTCCGACGAGTGGGCGCTCGGCGATGATGAGGCGGAGGGCGGCCGGGGGTTTTCACTCGCCGATAAGTTGGAGAGTCAAAGATTCGAGCACACGGACGCAGTCCTGGAGATGCAGGGCACCGATCTCACTGTCACATACTTGCAGAAGAATGGTTTTACAACTCCCTTGCTGTTCAAAGAGAAAACTGGTCTAGGATTGAGAGTACCGACGAGTAATTTCACTGTGAACGATGTGCGCATGTGCGTCGGTTCGCGCCGTATTCTCGACGTAATGGATGTCAACACACAGAAAAACATCGAGATGACCATGAAGGATTGGCAGCGTTACTATGACGATCCGAACAAAGAGCGTCTCTTGAACGTAATCTCTCTGGAGTTTTCGCACACTCGCCTCGAGAACTACGTGCAAGCTCCGCGTATTGTACGCCTGATAGATTGGGTCGATACCGTCTGGCCTCGACATTTGAAGGATCAACAGACTGAATCTACAAACGCACTCGATGACATGATGTACCCAAAAGTCCAGAAGTACTGCCTTATGTCTGTGAAGGGATGTTACACAGATTTCCACATTGATTTTGGCGGGACTTCCGTCTGGTATCACATTTTGAGAGGTGCAAAGGTGTTTTGGCTTATTCCACCAACTGAAAAGAATCTGCAACTCTATGAAAAGTGGGTTCTGTCCGGCAAGCAGTCTGATGTTTTCTTTGGTGACACAGTCGAGAAATGCATCAGAGTTCATCTGCAAGCGGGCCACACCTTCTTTATTCCGACGGGATGGATACACGCGGTGTACACTCCGAGCGACTCCCTTGTGTTTGGTGGAAATTTCCTTCATCAGTTTGGCATTGAAAAACAGTTGAAAATCGCACAAGTCGAAGACGTAACTAAGGTCCCACAAAAGTTCAGATACCCATTCTTCACGGAGATGCTGTGGTACGTGCTGGACCGGTACGTGTCCGCGTTGCTGGGTCGCTCGCACCTCGCGCCAGAGTGTCAGGCCTTGCCTCTCAACGCGCCCTCTGAACATGTTCACCTTACACAGAATGAACTGCACGGACTCAAGGCTATAGTTATGTATCTGCATCAGCTACCAGCGGCTCGCAAGTCGGTACCCGAATTACTCTCGGATCCCATTGCTCTGGTGAAAGACGTTCGCACTCTGGTCGAACAACATCGTCATGATAAACAGCACTTGGCCGTTACTGGAGTTCCAGTACTCAAAGGTCCAGAGGAGCCTCCCCTGACAGAGCGTCGCGGTGGTGGTGGGGGGCGCGGGGGGCGCGGCGGTGGGAGGGGAGGCGCGCGGCCACGCCCCGAACACGCCAACTCTGCTCCGCGACGCCGAAGGACTAGGTGTAAGAAGTGCGAGGCTTGTCAACGTACCGATTGTGGAGAATGCGTCTTTTGTCATGATATGGTTAAGTTCGGTGGTCAAGGACGTGCAAAACAAACGTGTATCATGCGACAGTGTCTACAGCCGATGTTACCAGTGACGGCTTCCTGTGCGGTGTGTCATTTGGACGGCTGGATGCAAACGCCGGTTGCGCCCCAAGCAAAGGGTAACTCCGGTCGTAACGGTCCGTCGTCTCTGCGCGAGTGCTCGGTGTGCTACAGCATCGTGCATCCGGCCTGCGTGCCCCCCGGCGGACAACTCAACGAGGATCTGCCCAACTCCTGGGAGTGTCCCACTTGCACCACCATGGGACTCAACCACGATTACAAGCCTCGACACTTTCGAGCTCGGCAAAAGTCATCAGACATACGCCGGATGAGTGTCAGCTCCGACGCCAGTCAACTCAGTCATCACGGAGTGCAGAACAAGCTGCCACCGCCGCTAACTCCCGTCACAACGAAGATAGATGGAGGCTCTGATAACGAAGCCAAGAATGACGTCCCACACATCAAATCGGAAGAGGATATAAAGCGGGAAGAGGAAGAAGAAAATTGCGTGGGCGGGGAAGAATCCGGTTCATGGGAGCCGCCGGGCAAAAAACGTCGCTCTTCTGATGACGATGAAGCGCCCAAGAAACAAGCGTTACGGGCTCACCTGGCCTTGCAGCTCACCCATCACACCGCCAAGTTATTGAAGAAGCCGATGTACCCGGTTCGTCCTGCGCTCGCGGCCGGCACCGCGGGAGGCTCGGGCTCGCCGTGGTTGGATCGAGAGGCGATGCTCTGTGTCTTCGCGAAGTTGACACCGCACGAGCTGGCCACTTGTGCGCTCGTCTGTAAAGCCTGGGCCGAATATTCGATGGATCCGTCCTTGTGGCGCAGCATGTCGTTTGTACAAGTGCGCGTCTCGGCGGCGCAGTTGGCCGGGATCGCTCGTCGACAGCCGCTCTCGCTCGGCCTAGAGTGGTGCCATCTCGCCAGGAGACAACTGGCTTGGTTGTTAGCCCGGCTGCCTGCACTGCGCTCTCTGTCGCTGGCCGGGTCCGCAGCTGAGGCCGCGCTCGCACTGCGCTCGGCGAGCTGCCCCCCTCTGCGGGCGCTGGACCTCTCGTTTGCTCGCTGCCTCGACGACGCTAAGCTGCGCGGGATACTGGCGCCCCCGGAGAACTCGCGGCCGGGGGGCGGAGCCCGAGCCGGGGAGCCCAGTCGCCTGGCCGCACTCGCTGTACTGCGTCTGCCCGGAACTGACATCACAGATGTAGCAATGAGATATGTTGTACAGACACTACCCAAACTGGTGGAACTGGATGCGTCGTCGTGCGCCCGACTCACGGACGCAGGTGCGGCGCAGTTAGCGCATCACTCGCTACAACGACTCTCGCTGGCCGGCTGCCGGCTGCTCACGGAGGCTGCTTTAGATCATCTCGCGCGCTGCCCCAATCTTGTTAGATTGGATCTCCGACATGTCCCTCTCGTATCGACACAGGCTGTGATTAAGTTTGCTGCCAAATCCAAACATAACCTTCACGTTAAGGATGTTAAGCTGGTTGAACTGCGGAAAACCTGA
Protein
MSETLPNNKKQVPRQLRERKHRKLYSDEWALGDDEAEGGRGFSLADKLESQRFEHTDAVLEMQGTDLTVTYLQKNGFTTPLLFKEKTGLGLRVPTSNFTVNDVRMCVGSRRILDVMDVNTQKNIEMTMKDWQRYYDDPNKERLLNVISLEFSHTRLENYVQAPRIVRLIDWVDTVWPRHLKDQQTESTNALDDMMYPKVQKYCLMSVKGCYTDFHIDFGGTSVWYHILRGAKVFWLIPPTEKNLQLYEKWVLSGKQSDVFFGDTVEKCIRVHLQAGHTFFIPTGWIHAVYTPSDSLVFGGNFLHQFGIEKQLKIAQVEDVTKVPQKFRYPFFTEMLWYVLDRYVSALLGRSHLAPECQALPLNAPSEHVHLTQNELHGLKAIVMYLHQLPAARKSVPELLSDPIALVKDVRTLVEQHRHDKQHLAVTGVPVLKGPEEPPLTERRGGGGGRGGRGGGRGGARPRPEHANSAPRRRRTRCKKCEACQRTDCGECVFCHDMVKFGGQGRAKQTCIMRQCLQPMLPVTASCAVCHLDGWMQTPVAPQAKGNSGRNGPSSLRECSVCYSIVHPACVPPGGQLNEDLPNSWECPTCTTMGLNHDYKPRHFRARQKSSDIRRMSVSSDASQLSHHGVQNKLPPPLTPVTTKIDGGSDNEAKNDVPHIKSEEDIKREEEEENCVGGEESGSWEPPGKKRRSSDDDEAPKKQALRAHLALQLTHHTAKLLKKPMYPVRPALAAGTAGGSGSPWLDREAMLCVFAKLTPHELATCALVCKAWAEYSMDPSLWRSMSFVQVRVSAAQLAGIARRQPLSLGLEWCHLARRQLAWLLARLPALRSLSLAGSAAEAALALRSASCPPLRALDLSFARCLDDAKLRGILAPPENSRPGGGARAGEPSRLAALAVLRLPGTDITDVAMRYVVQTLPKLVELDASSCARLTDAGAAQLAHHSLQRLSLAGCRLLTEAALDHLARCPNLVRLDLRHVPLVSTQAVIKFAAKSKHNLHVKDVKLVELRKT
Summary
Uniprot
EMBL
RSAL01000051
RVE50276.1
LADI01006439
KPJ20631.1
AGBW02007651
OWR54991.1
+ More
BABH01003764 BABH01003765 KQ460208 KPJ16599.1 ODYU01002363 SOQ39770.1 GFDF01003196 JAV10888.1 UFQS01000046 UFQT01000046 SSW98493.1 SSX18879.1 CVRI01000051 CRK99580.1 KQ971345 KYB27044.1 GECU01026914 JAS80792.1 GEDC01027261 JAS10037.1 APGK01019170 KB740092 KB632310 ENN81476.1 ERL92099.1 GL451753 EFN78650.1 GECU01002850 JAT04857.1
BABH01003764 BABH01003765 KQ460208 KPJ16599.1 ODYU01002363 SOQ39770.1 GFDF01003196 JAV10888.1 UFQS01000046 UFQT01000046 SSW98493.1 SSX18879.1 CVRI01000051 CRK99580.1 KQ971345 KYB27044.1 GECU01026914 JAS80792.1 GEDC01027261 JAS10037.1 APGK01019170 KB740092 KB632310 ENN81476.1 ERL92099.1 GL451753 EFN78650.1 GECU01002850 JAT04857.1
Proteomes
PRIDE
Pfam
Interpro
IPR041667
Cupin_8
+ More
IPR032675 LRR_dom_sf
IPR036047 F-box-like_dom_sf
IPR006553 Leu-rich_rpt_Cys-con_subtyp
IPR001965 Znf_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR019787 Znf_PHD-finger
IPR001810 F-box_dom
IPR041070 JHD
IPR003347 JmjC_dom
IPR002857 Znf_CXXC
IPR019786 Zinc_finger_PHD-type_CS
IPR001611 Leu-rich_rpt
IPR032675 LRR_dom_sf
IPR036047 F-box-like_dom_sf
IPR006553 Leu-rich_rpt_Cys-con_subtyp
IPR001965 Znf_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR019787 Znf_PHD-finger
IPR001810 F-box_dom
IPR041070 JHD
IPR003347 JmjC_dom
IPR002857 Znf_CXXC
IPR019786 Zinc_finger_PHD-type_CS
IPR001611 Leu-rich_rpt
SUPFAM
SSF81383
SSF81383
Gene 3D
ProteinModelPortal
PDB
2YU2
E-value=2.24637e-147,
Score=1343
Ontologies
GO
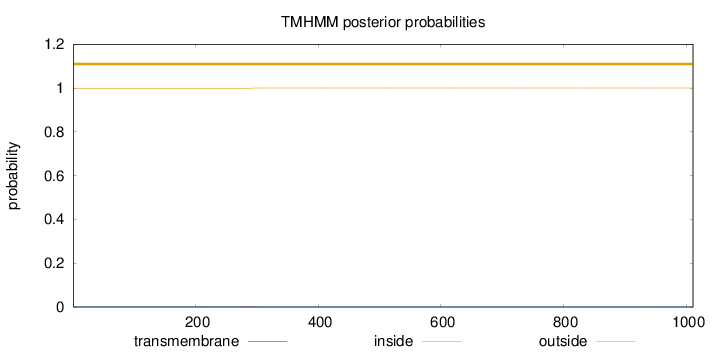
Topology
Length:
1011
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02867
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00125
outside
1 - 1011
Population Genetic Test Statistics
Pi
232.182845
Theta
174.136147
Tajima's D
1.109034
CLR
0.430347
CSRT
0.695815209239538
Interpretation
Uncertain
