Gene
KWMTBOMO09601
Pre Gene Modal
BGIBMGA012991
Annotation
PREDICTED:_calcium_release-activated_calcium_channel_protein_1-like_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.608
Sequence
CDS
ATGACTGTTACAACAGATGTTATTGCTTGGGACATTTGTTGGCTTGGGTACATTATCGGCCGAGCGCGGAAACTTCAATCTCGGCGTGCCGTGCTCGAGGCGTGGTCTCGCCGCCTGGCGGACCCCACCTACGGGCGACGGACTGTCGAGGCGATCCGCCCGGTCCTCTCGGACTGGGTGAATCGCGACCGAGGACGTCTCACCTTCCGAGCGACGCAGGTGCTCACGGGACACGGCTGTTTCGGTCGCTACCTGCACCTCGTCGCCCGGAGGGAGCCGACGCCGAAGTGTCACCACTGCAGTGGCTGCAACGAGGACACGGCGGAGCACACGCTCGCGTACTGCACCGCTTTCGCGGAGCAGCGCCGCGTCCTCGTTGCAAAAATAGGACCGGACTTGTCGCTTCCGACCGTCGTGGCTACGATGCTCGGCAGCGACGAGTCCTGGCAGGCGATGCTCGACTTCTGCGAGTCCACCATCTCGCAGAAGGAGGCGGCGGAACGGGAGAGGGAGAGCTCTTCTTCCCTCTCGGCGCCGTGCCGCCGCCGTCGAGCCGGGGTTCGGAGGAGGGCGTACTGTGGAGACCAGCCTAAAGACCTTCGCAACCGTCGAATTATGATAGCCGGCGTAATCAATATGTTGGAGGTACCAGACAATGTTTCGGAGGCGAGCTTCGTGACAAGCGTGGAGGAGTCTTGTCGAGTTGACCAATGTACTCAGACCCCACCGAGCGGAAAAAGCAGGCCACGCACCTTACAAAGGTATCACACTTCTGACCACATCTACGAGAGATCTGCGTACAGGAAACACAAGCACGAGCACTACCAGTTAATGACAAAGAGCCCAAGCTATGATGGACATCCGAAATTTAAAGTAAGCGTGACGGACAGCCCCACGATCTCCCTAACGCCTCCACATAGCTTGGAGTACGTTTGCCCTCGTGGGGGTACCAGCCCCAACGGCAGTACCACACCGAACGGGTTCTCGAACAAGTCTGCAGCCACCGTGCTACTGAATCCTAGTTATCTGCAGAACCCCTGTCAATGTAACCAGTCTATAAAGAGCATCAGTGAAGTGGCATTAGCTATGCCTGTCGCAAACTGGAACGAGCTTCCGGCTGACACGATCGGCTCCGAGGGCCTGTCGTGGCGAAGACTTCACATGTCCAGAGCCAAGCTCAAGGCCACGGCTACTACATCAGAATTATTATCAGGATTCGCTATGGTCGCAATGGTGGAACTACAAATCAACGAGCCGACGAATGTTCCCGAATGGCTGTTCGTGATGTTCGCCGTTTGCACAACGGTGCTCGTCGCTGTGCATATATTCGCTTTAATGATCTCTACATATTTATTGCCTAACATCGATGCAGTGAGCAAAATGGAAACGCCCGGTGGTCCTACAAGAGCTCTGAGGGATTCACCTCATGAGCGAATGAGGGGTTTTATAGAACTGGCTTGGGCATTTAGTACAGTTTTAGGTTTGTTCTTATTTTTGGTGGAAATTGCAATACTCTGCTGGGTGAAGTTTTGGGATTATTCATTTGCTGCTGCAACAGCAGCAACCGTAATTGTTATACCAGTGTTAATAGTTTTCGTGGCGTTCGCGATACATTTCTATCATTCGCTCGTTGTACAGAAGTGTGAGACCACCGTCAAAGACATTGAACAGCTCGAAAATATGAAGAGAGATCTCGACACGGCAACTGTCAAAGTGAACATGTACAACTGA
Protein
MTVTTDVIAWDICWLGYIIGRARKLQSRRAVLEAWSRRLADPTYGRRTVEAIRPVLSDWVNRDRGRLTFRATQVLTGHGCFGRYLHLVARREPTPKCHHCSGCNEDTAEHTLAYCTAFAEQRRVLVAKIGPDLSLPTVVATMLGSDESWQAMLDFCESTISQKEAAERERESSSSLSAPCRRRRAGVRRRAYCGDQPKDLRNRRIMIAGVINMLEVPDNVSEASFVTSVEESCRVDQCTQTPPSGKSRPRTLQRYHTSDHIYERSAYRKHKHEHYQLMTKSPSYDGHPKFKVSVTDSPTISLTPPHSLEYVCPRGGTSPNGSTTPNGFSNKSAATVLLNPSYLQNPCQCNQSIKSISEVALAMPVANWNELPADTIGSEGLSWRRLHMSRAKLKATATTSELLSGFAMVAMVELQINEPTNVPEWLFVMFAVCTTVLVAVHIFALMISTYLLPNIDAVSKMETPGGPTRALRDSPHERMRGFIELAWAFSTVLGLFLFLVEIAILCWVKFWDYSFAAATAATVIVIPVLIVFVAFAIHFYHSLVVQKCETTVKDIEQLENMKRDLDTATVKVNMYN
Summary
Uniprot
ProteinModelPortal
PDB
4HKR
E-value=1.93201e-50,
Score=505
Ontologies
GO
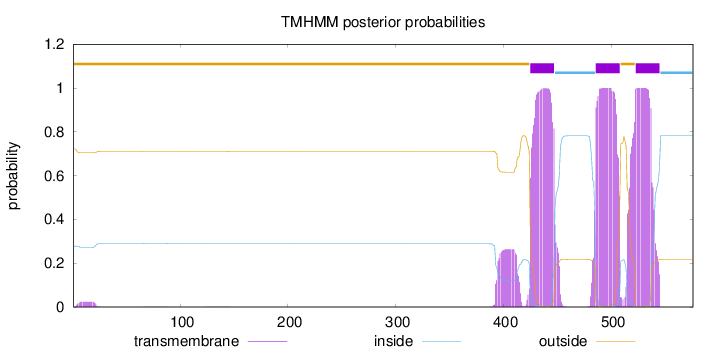
Topology
Length:
576
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
74.206
Exp number, first 60 AAs:
0.4118
Total prob of N-in:
0.27859
outside
1 - 424
TMhelix
425 - 447
inside
448 - 485
TMhelix
486 - 508
outside
509 - 522
TMhelix
523 - 545
inside
546 - 576
Population Genetic Test Statistics
Pi
237.104256
Theta
178.480543
Tajima's D
0.712619
CLR
0.467819
CSRT
0.575721213939303
Interpretation
Uncertain
