Pre Gene Modal
BGIBMGA014202
Annotation
PREDICTED:_NADH_dehydrogenase_[ubiquinone]_1_alpha_subcomplex_subunit_7-like_[Plutella_xylostella]
Location in the cell
Mitochondrial Reliability : 2.709
Sequence
CDS
ATGGTAAAGGCTAAAGTTCCACTACGCGACATTTCACCTGCATTGCAGGCTTTCAGGAACTTCCTCCTTGGTAGGAAGCATACAAACGCTCTTCGTTTTGAGCCATTGGTTTCGGCTAGGACCCAGCCTCCTCCAGAAATCCCAGACGGACCCTCGCACAAGCATGCCCACAACTACTATTACACAAGAGACGCTAGACGCGAAGTTGCTCCTCCAATTGATGTTACCAAGGAACTACTTAGTGCTAGCTCAGACAAAGGGGCTCCTAAACAAGCTGCTAATGTGAGACCTACACCTGGACACCTTTATGCTTGGGATAAACATTATGAATAA
Protein
MVKAKVPLRDISPALQAFRNFLLGRKHTNALRFEPLVSARTQPPPEIPDGPSHKHAHNYYYTRDARREVAPPIDVTKELLSASSDKGAPKQAANVRPTPGHLYAWDKHYE
Summary
Similarity
Belongs to the cytochrome P450 family.
Uniprot
H9JXD7
A0A2H1V8C9
A0A437BIJ5
A0A2A4J291
A0A0K8TBU5
S4PGZ4
+ More
A0A1E1WB74 A0A212FMN3 A0A2Z5U7E2 I4DJE4 A0A0N0PC52 I4DN45 A0A2M3Z6G2 A0A2M3ZI00 W5JJW8 A0A2M4C4J9 T1E9P8 A0A084VBV7 T1E3C6 A0A2M4AIK4 A0A182FNP3 A0A182V0D7 A0A182WYG9 Q7PTI2 A0A182I8I4 A0A182RBW3 A0A182VQD3 A0A182QC72 A0A182N6V9 A0A182XX94 B0X2B1 A0A1Q3FPZ0 A0A182KHF7 A0A1L8DDG7 A0A023ECR3 B4L2H3 A0A1B0DBL9 Q16QW8 U5EQJ5 A0A3B0JVQ2 Q29JM2 B4H386 B4M7W7 A0A182JF15 A0A3S6AAY9 B3P8X3 A0A0M4EUY8 O97418 A0A182SMN1 A0A1W4VRZ2 A0A1A9X807 A0A1B0ALU9 A0A1A9W2A1 A0A2J7RFF7 A0A2P8XE35 Q8SYB3 B4JXE1 A0A1A9ZLJ4 A0A1J1II85 D6WJ71 B4NQ19 A0A0L0BXW1 A0A2M3Z794 B4Q1A1 A0A336N0M8 A0A3Q7YK56 A0A182MSD2 A0A1B0GAH7 B4R3F1 B4I9Q6 A0A067QTT3 A0A2M4CL85 B3MYR4 A0A1A9UU65 A0A0K8TTG3 T1HZC8 A0A069DMJ9 A0A0A9WDX7 A0A1B6CYH8 A0A1B6JE77 A0A1I8P893 A0A1W4WHA6 A0A1I8MCP2 A0A1Y1NI93 A0A0V0G717 A0A1L8EEK0 A0A1B6L428 A0A1L8EEW7 A0A162R1H7 A0A0N8C338 A0A0N8ASI7 N6TPQ3 A0A0P6BI03 A0A0N8ANK4 A0A423U4V1 A0A194R1S5
A0A1E1WB74 A0A212FMN3 A0A2Z5U7E2 I4DJE4 A0A0N0PC52 I4DN45 A0A2M3Z6G2 A0A2M3ZI00 W5JJW8 A0A2M4C4J9 T1E9P8 A0A084VBV7 T1E3C6 A0A2M4AIK4 A0A182FNP3 A0A182V0D7 A0A182WYG9 Q7PTI2 A0A182I8I4 A0A182RBW3 A0A182VQD3 A0A182QC72 A0A182N6V9 A0A182XX94 B0X2B1 A0A1Q3FPZ0 A0A182KHF7 A0A1L8DDG7 A0A023ECR3 B4L2H3 A0A1B0DBL9 Q16QW8 U5EQJ5 A0A3B0JVQ2 Q29JM2 B4H386 B4M7W7 A0A182JF15 A0A3S6AAY9 B3P8X3 A0A0M4EUY8 O97418 A0A182SMN1 A0A1W4VRZ2 A0A1A9X807 A0A1B0ALU9 A0A1A9W2A1 A0A2J7RFF7 A0A2P8XE35 Q8SYB3 B4JXE1 A0A1A9ZLJ4 A0A1J1II85 D6WJ71 B4NQ19 A0A0L0BXW1 A0A2M3Z794 B4Q1A1 A0A336N0M8 A0A3Q7YK56 A0A182MSD2 A0A1B0GAH7 B4R3F1 B4I9Q6 A0A067QTT3 A0A2M4CL85 B3MYR4 A0A1A9UU65 A0A0K8TTG3 T1HZC8 A0A069DMJ9 A0A0A9WDX7 A0A1B6CYH8 A0A1B6JE77 A0A1I8P893 A0A1W4WHA6 A0A1I8MCP2 A0A1Y1NI93 A0A0V0G717 A0A1L8EEK0 A0A1B6L428 A0A1L8EEW7 A0A162R1H7 A0A0N8C338 A0A0N8ASI7 N6TPQ3 A0A0P6BI03 A0A0N8ANK4 A0A423U4V1 A0A194R1S5
Pubmed
19121390
23622113
22118469
22651552
26354079
20920257
+ More
23761445 24438588 24330624 9087549 12364791 14747013 17210077 25244985 24945155 17994087 17510324 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 18362917 19820115 26108605 17550304 24845553 26369729 26334808 25401762 26823975 25315136 28004739 23537049
23761445 24438588 24330624 9087549 12364791 14747013 17210077 25244985 24945155 17994087 17510324 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 18362917 19820115 26108605 17550304 24845553 26369729 26334808 25401762 26823975 25315136 28004739 23537049
EMBL
BABH01042617
ODYU01001204
SOQ37103.1
RSAL01000051
RVE50256.1
NWSH01004073
+ More
PCG65512.1 GBRD01003186 JAG62635.1 GAIX01005995 JAA86565.1 GDQN01006838 JAT84216.1 AGBW02007651 OWR54997.1 AP017524 BBB06879.1 AK401412 BAM18034.1 KQ460773 KPJ12442.1 AK402713 BAM19335.1 GGFM01003333 MBW24084.1 GGFM01007319 MBW28070.1 ADMH02001019 ETN64396.1 GGFJ01011044 MBW60185.1 GAMD01001137 JAB00454.1 ATLV01009559 KE524536 KFB35451.1 GALA01000480 JAA94372.1 GGFK01007256 MBW40577.1 AAAB01007888 AAAB01008805 EAA03125.3 EAA03888.3 APCN01003034 AXCN02000309 DS232284 EDS39134.1 GFDL01005473 JAV29572.1 GFDF01009704 JAV04380.1 GAPW01006451 GEHC01000210 JAC07147.1 JAV47435.1 CH933810 EDW06849.1 AJVK01013711 CH477729 EAT36783.1 GANO01004250 JAB55621.1 OUUW01000003 SPP77799.1 CH379063 EAL32279.1 CH479205 EDW30779.1 CH940653 EDW62884.1 GQ314895 ADF67663.1 CH954183 EDV45578.1 CP012528 ALC48985.1 AE014298 BT056316 AL009194 AAF45739.1 ACL68763.1 AHN59276.1 CAA15701.1 JXJN01000159 NEVH01004413 PNF39571.1 PYGN01002564 PSN30267.1 AY071663 AAL49285.1 CH916376 EDV95417.1 CVRI01000051 CRK99482.1 KQ971343 EFA04705.1 CH964291 EDW86244.1 JRES01001169 KNC24831.1 GGFM01003609 MBW24360.1 GQ314872 GQ314873 GQ314874 GQ314875 GQ314876 GQ314877 GQ314878 GQ314879 GQ314880 GQ314881 GQ314882 GQ314883 GQ314884 GQ314885 CM000162 ADF67640.1 ADF67641.1 ADF67642.1 ADF67643.1 ADF67644.1 ADF67645.1 ADF67646.1 ADF67647.1 ADF67648.1 ADF67649.1 ADF67650.1 ADF67651.1 ADF67652.1 ADF67653.1 EDX01408.1 UFQS01000056 UFQT01000056 UFQT01005558 SSW98748.1 SSX36026.1 GQ314886 GQ314887 GQ314888 GQ314889 GQ314890 GQ314891 GQ314892 GQ314893 GQ314894 GQ314896 GQ314897 GQ314898 ADF67654.1 ADF67655.1 ADF67656.1 ADF67657.1 ADF67658.1 ADF67659.1 ADF67660.1 ADF67661.1 ADF67662.1 ADF67664.1 ADF67665.1 ADF67666.1 AXCM01002699 CCAG010017803 CM000366 EDX16903.1 CH480825 EDW43937.1 KK853474 KDR07358.1 GGFL01001753 MBW65931.1 CH902632 EDV32758.1 GDAI01000157 JAI17446.1 ACPB03016118 GBGD01003611 JAC85278.1 GBHO01040534 GBRD01009913 GDHC01014527 JAG03070.1 JAG55911.1 JAQ04102.1 GEDC01018913 JAS18385.1 GECU01010298 JAS97408.1 GEZM01005999 JAV95836.1 GECL01002389 JAP03735.1 GFDG01001657 JAV17142.1 GEBQ01021606 JAT18371.1 GFDG01001660 JAV17139.1 LRGB01000243 KZS20144.1 GDIQ01119555 JAL32171.1 GDIQ01247163 JAK04562.1 APGK01059435 KB741293 KB632046 ENN70266.1 ERL88275.1 GDIP01022097 JAM81618.1 GDIQ01258227 JAJ93497.1 QCYY01000643 ROT83702.1 KQ460883 KPJ11200.1
PCG65512.1 GBRD01003186 JAG62635.1 GAIX01005995 JAA86565.1 GDQN01006838 JAT84216.1 AGBW02007651 OWR54997.1 AP017524 BBB06879.1 AK401412 BAM18034.1 KQ460773 KPJ12442.1 AK402713 BAM19335.1 GGFM01003333 MBW24084.1 GGFM01007319 MBW28070.1 ADMH02001019 ETN64396.1 GGFJ01011044 MBW60185.1 GAMD01001137 JAB00454.1 ATLV01009559 KE524536 KFB35451.1 GALA01000480 JAA94372.1 GGFK01007256 MBW40577.1 AAAB01007888 AAAB01008805 EAA03125.3 EAA03888.3 APCN01003034 AXCN02000309 DS232284 EDS39134.1 GFDL01005473 JAV29572.1 GFDF01009704 JAV04380.1 GAPW01006451 GEHC01000210 JAC07147.1 JAV47435.1 CH933810 EDW06849.1 AJVK01013711 CH477729 EAT36783.1 GANO01004250 JAB55621.1 OUUW01000003 SPP77799.1 CH379063 EAL32279.1 CH479205 EDW30779.1 CH940653 EDW62884.1 GQ314895 ADF67663.1 CH954183 EDV45578.1 CP012528 ALC48985.1 AE014298 BT056316 AL009194 AAF45739.1 ACL68763.1 AHN59276.1 CAA15701.1 JXJN01000159 NEVH01004413 PNF39571.1 PYGN01002564 PSN30267.1 AY071663 AAL49285.1 CH916376 EDV95417.1 CVRI01000051 CRK99482.1 KQ971343 EFA04705.1 CH964291 EDW86244.1 JRES01001169 KNC24831.1 GGFM01003609 MBW24360.1 GQ314872 GQ314873 GQ314874 GQ314875 GQ314876 GQ314877 GQ314878 GQ314879 GQ314880 GQ314881 GQ314882 GQ314883 GQ314884 GQ314885 CM000162 ADF67640.1 ADF67641.1 ADF67642.1 ADF67643.1 ADF67644.1 ADF67645.1 ADF67646.1 ADF67647.1 ADF67648.1 ADF67649.1 ADF67650.1 ADF67651.1 ADF67652.1 ADF67653.1 EDX01408.1 UFQS01000056 UFQT01000056 UFQT01005558 SSW98748.1 SSX36026.1 GQ314886 GQ314887 GQ314888 GQ314889 GQ314890 GQ314891 GQ314892 GQ314893 GQ314894 GQ314896 GQ314897 GQ314898 ADF67654.1 ADF67655.1 ADF67656.1 ADF67657.1 ADF67658.1 ADF67659.1 ADF67660.1 ADF67661.1 ADF67662.1 ADF67664.1 ADF67665.1 ADF67666.1 AXCM01002699 CCAG010017803 CM000366 EDX16903.1 CH480825 EDW43937.1 KK853474 KDR07358.1 GGFL01001753 MBW65931.1 CH902632 EDV32758.1 GDAI01000157 JAI17446.1 ACPB03016118 GBGD01003611 JAC85278.1 GBHO01040534 GBRD01009913 GDHC01014527 JAG03070.1 JAG55911.1 JAQ04102.1 GEDC01018913 JAS18385.1 GECU01010298 JAS97408.1 GEZM01005999 JAV95836.1 GECL01002389 JAP03735.1 GFDG01001657 JAV17142.1 GEBQ01021606 JAT18371.1 GFDG01001660 JAV17139.1 LRGB01000243 KZS20144.1 GDIQ01119555 JAL32171.1 GDIQ01247163 JAK04562.1 APGK01059435 KB741293 KB632046 ENN70266.1 ERL88275.1 GDIP01022097 JAM81618.1 GDIQ01258227 JAJ93497.1 QCYY01000643 ROT83702.1 KQ460883 KPJ11200.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053240
UP000000673
+ More
UP000030765 UP000069272 UP000075903 UP000076407 UP000007062 UP000075840 UP000075900 UP000075920 UP000075886 UP000075884 UP000076408 UP000002320 UP000075881 UP000009192 UP000092462 UP000008820 UP000268350 UP000001819 UP000008744 UP000008792 UP000075880 UP000008711 UP000092553 UP000000803 UP000075901 UP000192221 UP000092443 UP000092460 UP000091820 UP000235965 UP000245037 UP000001070 UP000092445 UP000183832 UP000007266 UP000007798 UP000037069 UP000002282 UP000075883 UP000092444 UP000000304 UP000001292 UP000027135 UP000007801 UP000078200 UP000015103 UP000095300 UP000192223 UP000095301 UP000076858 UP000019118 UP000030742 UP000283509
UP000030765 UP000069272 UP000075903 UP000076407 UP000007062 UP000075840 UP000075900 UP000075920 UP000075886 UP000075884 UP000076408 UP000002320 UP000075881 UP000009192 UP000092462 UP000008820 UP000268350 UP000001819 UP000008744 UP000008792 UP000075880 UP000008711 UP000092553 UP000000803 UP000075901 UP000192221 UP000092443 UP000092460 UP000091820 UP000235965 UP000245037 UP000001070 UP000092445 UP000183832 UP000007266 UP000007798 UP000037069 UP000002282 UP000075883 UP000092444 UP000000304 UP000001292 UP000027135 UP000007801 UP000078200 UP000015103 UP000095300 UP000192223 UP000095301 UP000076858 UP000019118 UP000030742 UP000283509
Interpro
Gene 3D
ProteinModelPortal
H9JXD7
A0A2H1V8C9
A0A437BIJ5
A0A2A4J291
A0A0K8TBU5
S4PGZ4
+ More
A0A1E1WB74 A0A212FMN3 A0A2Z5U7E2 I4DJE4 A0A0N0PC52 I4DN45 A0A2M3Z6G2 A0A2M3ZI00 W5JJW8 A0A2M4C4J9 T1E9P8 A0A084VBV7 T1E3C6 A0A2M4AIK4 A0A182FNP3 A0A182V0D7 A0A182WYG9 Q7PTI2 A0A182I8I4 A0A182RBW3 A0A182VQD3 A0A182QC72 A0A182N6V9 A0A182XX94 B0X2B1 A0A1Q3FPZ0 A0A182KHF7 A0A1L8DDG7 A0A023ECR3 B4L2H3 A0A1B0DBL9 Q16QW8 U5EQJ5 A0A3B0JVQ2 Q29JM2 B4H386 B4M7W7 A0A182JF15 A0A3S6AAY9 B3P8X3 A0A0M4EUY8 O97418 A0A182SMN1 A0A1W4VRZ2 A0A1A9X807 A0A1B0ALU9 A0A1A9W2A1 A0A2J7RFF7 A0A2P8XE35 Q8SYB3 B4JXE1 A0A1A9ZLJ4 A0A1J1II85 D6WJ71 B4NQ19 A0A0L0BXW1 A0A2M3Z794 B4Q1A1 A0A336N0M8 A0A3Q7YK56 A0A182MSD2 A0A1B0GAH7 B4R3F1 B4I9Q6 A0A067QTT3 A0A2M4CL85 B3MYR4 A0A1A9UU65 A0A0K8TTG3 T1HZC8 A0A069DMJ9 A0A0A9WDX7 A0A1B6CYH8 A0A1B6JE77 A0A1I8P893 A0A1W4WHA6 A0A1I8MCP2 A0A1Y1NI93 A0A0V0G717 A0A1L8EEK0 A0A1B6L428 A0A1L8EEW7 A0A162R1H7 A0A0N8C338 A0A0N8ASI7 N6TPQ3 A0A0P6BI03 A0A0N8ANK4 A0A423U4V1 A0A194R1S5
A0A1E1WB74 A0A212FMN3 A0A2Z5U7E2 I4DJE4 A0A0N0PC52 I4DN45 A0A2M3Z6G2 A0A2M3ZI00 W5JJW8 A0A2M4C4J9 T1E9P8 A0A084VBV7 T1E3C6 A0A2M4AIK4 A0A182FNP3 A0A182V0D7 A0A182WYG9 Q7PTI2 A0A182I8I4 A0A182RBW3 A0A182VQD3 A0A182QC72 A0A182N6V9 A0A182XX94 B0X2B1 A0A1Q3FPZ0 A0A182KHF7 A0A1L8DDG7 A0A023ECR3 B4L2H3 A0A1B0DBL9 Q16QW8 U5EQJ5 A0A3B0JVQ2 Q29JM2 B4H386 B4M7W7 A0A182JF15 A0A3S6AAY9 B3P8X3 A0A0M4EUY8 O97418 A0A182SMN1 A0A1W4VRZ2 A0A1A9X807 A0A1B0ALU9 A0A1A9W2A1 A0A2J7RFF7 A0A2P8XE35 Q8SYB3 B4JXE1 A0A1A9ZLJ4 A0A1J1II85 D6WJ71 B4NQ19 A0A0L0BXW1 A0A2M3Z794 B4Q1A1 A0A336N0M8 A0A3Q7YK56 A0A182MSD2 A0A1B0GAH7 B4R3F1 B4I9Q6 A0A067QTT3 A0A2M4CL85 B3MYR4 A0A1A9UU65 A0A0K8TTG3 T1HZC8 A0A069DMJ9 A0A0A9WDX7 A0A1B6CYH8 A0A1B6JE77 A0A1I8P893 A0A1W4WHA6 A0A1I8MCP2 A0A1Y1NI93 A0A0V0G717 A0A1L8EEK0 A0A1B6L428 A0A1L8EEW7 A0A162R1H7 A0A0N8C338 A0A0N8ASI7 N6TPQ3 A0A0P6BI03 A0A0N8ANK4 A0A423U4V1 A0A194R1S5
PDB
6G72
E-value=7.11269e-08,
Score=129
Ontologies
PATHWAY
GO
PANTHER
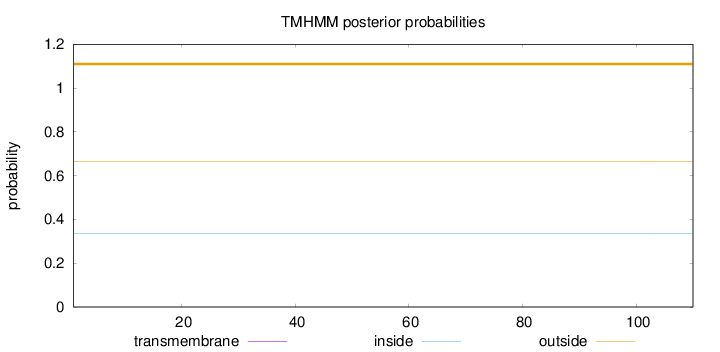
Topology
Length:
110
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00054
Exp number, first 60 AAs:
0.00054
Total prob of N-in:
0.33452
outside
1 - 110
Population Genetic Test Statistics
Pi
33.023595
Theta
33.876384
Tajima's D
-0.63097
CLR
0.440216
CSRT
0.210139493025349
Interpretation
Uncertain
