Gene
KWMTBOMO09597 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014201
Annotation
PREDICTED:_cytosolic_10-formyltetrahydrofolate_dehydrogenase_[Bombyx_mori]
Full name
10-formyltetrahydrofolate dehydrogenase
+ More
Cytosolic 10-formyltetrahydrofolate dehydrogenase
Cytosolic 10-formyltetrahydrofolate dehydrogenase
Alternative Name
Aldehyde dehydrogenase family 1 member L1
Location in the cell
Cytoplasmic Reliability : 3.407
Sequence
CDS
ATGCCGCCAGTATCATTGACTGATCATGCACCAAAAAAAAGCCTAAGAGTTGCCATTATTGGCCAAAGTACATTTGCAGCTGAAGTTTTCAAACTCATCCAAAGAGACGGTCATGAAGTTGTGGGTGTCTTCACAATTCCTGACAAAGGCAACAGGGAGGACCCTTTAGCTACTATTGCCGCACAGAATGGCAAACCAGTTTACAAATACAAGTCTTGGCGTATTAAAGGACAAGTCATCCCTGAAGTACTTGAGCAGTACAAATCGGTGAACGCCGACCTCAATGTTCTACCGTTCTGCACTCAATTCATACCCATGGAGGTGATCACGGCTCCTAAATACGAGAGCATCTGTTACCATCCGAGCATTTTACCCAGACATCGGGGAGCTTCATCAGTTAATTGGACCTTGATCGAGGGTGACACTACATGCGGCTTCTCCATATTTTGGGCGGACGACGGCTTAGACACCGGCCCCATATTACTGCAAAAGAGCTATCCCACAACCATCGATGACACCGTAGACACGATCTACAACAAATTCTTATATCCTGAAGGTATCAAAGCCTTAGCTGAAGCCGTCAACATGGTTGCGAATGGTGTGGCTCCTAAAATCAAGCAGTCTGAAGAAGGCGCCACATACGACCCAGCTCTTTTCAAACCTGAAACTCATCTGATCGATTGGAGCAAAGGAGGGCTGGCTCTGCACAACTTCGTGCGAGGACTGGACTCATCACCGGGAGCCACCTCCTTCATCGAACCTCAGACCAAAGAGGGGATAATCGGAGACGGTGAAAAAATTGAGATAAAATTCTTCGGATCCTCCCTCTGGCAATCTGAATATGAGACCGAAGGAGATAAACTGTATATAACAGGCCTTAATAAACCAGCCGTGGTCCACGAAGCCGGCTTACTTGTTACTGCCAATGATGGAGTTAGGCTCAACATACAGAGGCTAAAAGTTAACGGGAAGATGATCAACGCTCAAAACTTCTTCAAAGCTAGCGAAGAGAAGATCTCTATAGAACTGAATTCGGATGAGTTAAAGTTCGTGGAGAGCTGCCGCGAGGTGTGGAGGGCGATTCTTCGCATTGAAATCGAAAACCACACCGACTTCTTCGCATCCGGAGCCGGCTCTATGGACGTCGTGAGGCTGGTCGAAGAAGTCAAGGACTTGGCGAACGTGGAGCTCCAGAATGAGGATGTATACATGAACACCACGTTTGAAGAGTTCTATAGTGTGGCCGTGTTGAAGGCGAGAGGCGGCTCGGATACAAAAGAGGTTGTTTATGAGGGACTCGAAATGGAGGTCAATAAAATGAAGATTAAATTCCCGACACAGCTGTTTATTAATGGCGAGTTCGTGAACGCTGACAGTGGTAAAACCTTGACCCTGATCAATCCAACGGACGAGAGCGTGATCTGTAAAGTGCAAAGCGCCTCGCCCAACGACGTTGACAAGGCAGTGCGGGCCGCTAAGAAGGCGTTCGATGAAGGGGAATGGTCCAAGATCAGTGCCCGTGAGAGAGCACAAATCTTATTTAAGTTGGCAGATTTGATGGAACAACACAAAGAGGAGCTGGCCACAATCGAATCCATAGACTCGGGAGCTGTCTACACGTTAGCACTCAAAACCCACGTGGGCATGTCCATAGACACGTGGCGCTACTTCGCTGGCTGGTGCGACAAGATCCAAGGCGCCACCGTGCCCATAAACCACGCCAGGCCAAACAGAAATCTGTGCATCACGAGAAAAGAGCCGATCGGCGTTTGCGCGATAATAACGCCCTGGAATTATCCGCTGATGATGCTGTCGTGGAAGATGGCGGCCTGTCTCGCTGCTGGCAATACTGTCGTCATGAAACCCGCTGCGGTGTGCCCCCTGACAGCGTTGAAGTTCGCCGAGCTGTGCGTCCGAGCGGGGGTTCCCCCCGGCGTCGTGAACATCCTCCCCGGGAGCGGCGCGGCGTGCGGGCAGGCGCTCGCCGACCATCCGCTCGTCCGGAAGCTGGGCTTCACAGGCAGCACTCCCATCGGTCAAACCATAATGAAGTCGTGTGCAGCATCGAATATGAAGAAGGTGTCGTTGGAGCTAGGAGGCAAGTCGCCTCTCGTTATATTCGAAGACTGCGATCTGGACAAAGCCGTCAGAAACGGTATGGCAGCTGTGTTCTTCAATAAAGGCGAGAATTGCATAGCAGCAGGTCGGTTGTTCGTTGAAGAGACGATACACGACGAGTTCGTACGTCGCGTTATAGAGGAGACCGAGAAGATCCGTATCGGCGACCCACTGCACCGGGGCACTTCACACGGACCCCAGAACCATAAGGCGCATATGGATAAACTGATAGAGTACTGCGAGCGTGGCGTCAAAGAAGGTGCCAAGCTAGTGTACGGAGGCAAGAGGGTGGATAGAAAAGGCTATTTCTTCCAGCCGACTGTCTTTACCGACGTCACAGACGAGATGTACATCGCCAAGGAGGAATCGTTTGGACCCATCATGATCATCAGCAAGTTCAGCAGCAAGAATTTGGAGGACGTGATCCGTCGCGCGAACAACACGGAGTATGGGCTCGCGAGCGGCGTGTTCACGCGGGACGTGTCGCGGGCGCTGCAGTTCGCCGAGCGCATCGACGCGGGCACGGTGTTCGTCAACACGTACAACAAGACCGACGTGGCGGCGCCCTTCGGAGGCTTCAAGCAGTCCGGCTTCGGAAAGGACTTGGGCCAAGACGCTCTCAACGAATATCTGAAAACGAAATGCGTGACAATAGAATATTAA
Protein
MPPVSLTDHAPKKSLRVAIIGQSTFAAEVFKLIQRDGHEVVGVFTIPDKGNREDPLATIAAQNGKPVYKYKSWRIKGQVIPEVLEQYKSVNADLNVLPFCTQFIPMEVITAPKYESICYHPSILPRHRGASSVNWTLIEGDTTCGFSIFWADDGLDTGPILLQKSYPTTIDDTVDTIYNKFLYPEGIKALAEAVNMVANGVAPKIKQSEEGATYDPALFKPETHLIDWSKGGLALHNFVRGLDSSPGATSFIEPQTKEGIIGDGEKIEIKFFGSSLWQSEYETEGDKLYITGLNKPAVVHEAGLLVTANDGVRLNIQRLKVNGKMINAQNFFKASEEKISIELNSDELKFVESCREVWRAILRIEIENHTDFFASGAGSMDVVRLVEEVKDLANVELQNEDVYMNTTFEEFYSVAVLKARGGSDTKEVVYEGLEMEVNKMKIKFPTQLFINGEFVNADSGKTLTLINPTDESVICKVQSASPNDVDKAVRAAKKAFDEGEWSKISARERAQILFKLADLMEQHKEELATIESIDSGAVYTLALKTHVGMSIDTWRYFAGWCDKIQGATVPINHARPNRNLCITRKEPIGVCAIITPWNYPLMMLSWKMAACLAAGNTVVMKPAAVCPLTALKFAELCVRAGVPPGVVNILPGSGAACGQALADHPLVRKLGFTGSTPIGQTIMKSCAASNMKKVSLELGGKSPLVIFEDCDLDKAVRNGMAAVFFNKGENCIAAGRLFVEETIHDEFVRRVIEETEKIRIGDPLHRGTSHGPQNHKAHMDKLIEYCERGVKEGAKLVYGGKRVDRKGYFFQPTVFTDVTDEMYIAKEESFGPIMIISKFSSKNLEDVIRRANNTEYGLASGVFTRDVSRALQFAERIDAGTVFVNTYNKTDVAAPFGGFKQSGFGKDLGQDALNEYLKTKCVTIEY
Summary
Catalytic Activity
(6S)-10-formyltetrahydrofolate + H2O + NADP(+) = (6S)-5,6,7,8-tetrahydrofolate + CO2 + H(+) + NADPH
Subunit
Homotetramer.
Similarity
Belongs to the aldehyde dehydrogenase family.
In the C-terminal section; belongs to the aldehyde dehydrogenase family. ALDH1L subfamily.
In the N-terminal section; belongs to the GART family.
In the C-terminal section; belongs to the aldehyde dehydrogenase family. ALDH1L subfamily.
In the N-terminal section; belongs to the GART family.
Keywords
Complete proteome
Cytoplasm
NADP
One-carbon metabolism
Oxidoreductase
Phosphopantetheine
Phosphoprotein
Reference proteome
Feature
chain Cytosolic 10-formyltetrahydrofolate dehydrogenase
Uniprot
A0A2H1W8W8
A0A2A4J0X0
A0A212FMM3
A0A194PRS4
A0A437BIX8
A0A1L8D6P4
+ More
A0A194RER6 D2A5X9 A0A154PHP2 A0A411G7T9 A0A0M9A4E9 A0A2J7Q7B7 A0A067QH51 F4W7E1 A0A087ZYB7 A0A2L2Y5D2 A0A1Y1LFN3 A0A2L2Y5B9 A0A195DSU2 A0A151X8P0 A0A3R7PD39 A0A151JT46 A0A232FKK8 A0A151IHD6 A0A158NPV3 K7ILV9 A0A195ATU6 E2A3S4 A0A0L7R0K3 A0A026WU32 A0A3L8D917 A0A1Z5KX88 E9IC80 A0A0N8CK90 A0A1L8D6Q1 A0A0P5QMT0 A0A1B0GMU0 A0A0Q9X4L0 B4KEZ9 A0A1B0EY70 A0A3S3PF55 A0A2P2HYA6 T1JDA6 A0A1B6F6T2 A0A2H8TLQ5 A0A0K8WIE2 A0A0C9RMA6 W8BRG1 A0A3B0JV67 X1WIZ3 B4M9T3 A0A0M4EA78 A0A3Q1BVF1 Q63ZT8 A0A3P8TBZ4 E9H8V6 A0A0P5ZGX4 B4GT55 A0A443SUZ4 A0A3Q1HEV9 Q6GNL7 Q29KL6 A0A1S3IEW6 A0A1L8GPP8 A0A3Q2QVF3 A0A3B4Z686 A0A3Q1G9C9 A0A3B4U107 A0A146ZX33 A0A3P9QAE5 I3J595 A0A3P9BES5 B0WX46 A0A0P5QR58 B4Q4E1 A0A3B4XY61 A0A3B4FIR4 B3MMB2 B4N7Q7 A0A1W4VAS3 B4JPX0 B4IFN9 B3NLW9 A0A3Q2WDL4 Q16RZ0 A0A182MKG5 Q9VIC9 A0A3Q3NF04 Q6NNU7 A0A182WHJ5 A0A0P7YKU6 A0A0L0CLW3 A0A3Q2Z4G1 A0A1S3P7F6 A0A164XG39 A0A1W4Y042 A0A182RRH5 A0A182Q3G5 A0A2Z5UMA5 B4P6W5
A0A194RER6 D2A5X9 A0A154PHP2 A0A411G7T9 A0A0M9A4E9 A0A2J7Q7B7 A0A067QH51 F4W7E1 A0A087ZYB7 A0A2L2Y5D2 A0A1Y1LFN3 A0A2L2Y5B9 A0A195DSU2 A0A151X8P0 A0A3R7PD39 A0A151JT46 A0A232FKK8 A0A151IHD6 A0A158NPV3 K7ILV9 A0A195ATU6 E2A3S4 A0A0L7R0K3 A0A026WU32 A0A3L8D917 A0A1Z5KX88 E9IC80 A0A0N8CK90 A0A1L8D6Q1 A0A0P5QMT0 A0A1B0GMU0 A0A0Q9X4L0 B4KEZ9 A0A1B0EY70 A0A3S3PF55 A0A2P2HYA6 T1JDA6 A0A1B6F6T2 A0A2H8TLQ5 A0A0K8WIE2 A0A0C9RMA6 W8BRG1 A0A3B0JV67 X1WIZ3 B4M9T3 A0A0M4EA78 A0A3Q1BVF1 Q63ZT8 A0A3P8TBZ4 E9H8V6 A0A0P5ZGX4 B4GT55 A0A443SUZ4 A0A3Q1HEV9 Q6GNL7 Q29KL6 A0A1S3IEW6 A0A1L8GPP8 A0A3Q2QVF3 A0A3B4Z686 A0A3Q1G9C9 A0A3B4U107 A0A146ZX33 A0A3P9QAE5 I3J595 A0A3P9BES5 B0WX46 A0A0P5QR58 B4Q4E1 A0A3B4XY61 A0A3B4FIR4 B3MMB2 B4N7Q7 A0A1W4VAS3 B4JPX0 B4IFN9 B3NLW9 A0A3Q2WDL4 Q16RZ0 A0A182MKG5 Q9VIC9 A0A3Q3NF04 Q6NNU7 A0A182WHJ5 A0A0P7YKU6 A0A0L0CLW3 A0A3Q2Z4G1 A0A1S3P7F6 A0A164XG39 A0A1W4Y042 A0A182RRH5 A0A182Q3G5 A0A2Z5UMA5 B4P6W5
EC Number
1.5.1.6
Pubmed
22118469
26354079
18362917
19820115
24845553
21719571
+ More
26561354 28004739 28648823 21347285 20075255 20798317 24508170 30249741 28528879 21282665 17994087 24495485 21292972 15632085 27762356 25186727 22936249 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 17550304
26561354 28004739 28648823 21347285 20075255 20798317 24508170 30249741 28528879 21282665 17994087 24495485 21292972 15632085 27762356 25186727 22936249 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 17550304
EMBL
ODYU01007067
SOQ49510.1
NWSH01004073
PCG65509.1
AGBW02007651
OWR54998.1
+ More
KQ459595 KPI96012.1 RSAL01000051 RVE50258.1 GEYN01000007 JAV02122.1 KQ460323 KPJ15954.1 KQ971345 EFA05437.1 KQ434912 KZC11396.1 MH366154 QBB01963.1 KQ435760 KOX75639.1 NEVH01017446 PNF24469.1 KK853412 KDR07781.1 GL887844 EGI69814.1 IAAA01004539 LAA03364.1 GEZM01057134 JAV72434.1 IAAA01004538 LAA03361.1 KQ980487 KYN15913.1 KQ982409 KYQ56732.1 QCYY01002845 ROT67137.1 KQ982014 KYN30626.1 NNAY01000091 OXU31070.1 KQ977606 KYN01418.1 ADTU01022757 ADTU01022758 KQ976745 KYM75482.1 GL436457 EFN71966.1 KQ414670 KOC64382.1 KK107105 EZA59493.1 QOIP01000011 RLU16408.1 GFJQ02007217 JAV99752.1 GL762206 EFZ21820.1 GDIP01123527 JAL80187.1 GEYN01000008 JAV02121.1 GDIQ01112119 JAL39607.1 AJVK01003753 AJVK01003754 CH933807 KRG03008.1 EDW11968.2 AJWK01015367 NCKU01000824 RWS14003.1 IACF01001021 LAB66751.1 JH432102 GECZ01024196 JAS45573.1 GFXV01002797 MBW14602.1 GDHF01001500 JAI50814.1 GBYB01008111 GBYB01008114 JAG77878.1 JAG77881.1 GAMC01010719 JAB95836.1 OUUW01000010 SPP85985.1 ABLF02036745 ABLF02036754 ABLF02058045 ABLF02059108 CH940654 EDW57959.1 CP012523 ALC39251.1 CR926185 CR926221 BC082822 GL732606 EFX71787.1 GDIP01043744 JAM59971.1 CH479189 EDW25564.1 NCKV01000190 RWS31346.1 BC073490 CH379061 EAL33158.1 CM004472 OCT85802.1 GCES01015798 JAR70525.1 AERX01016021 AERX01016022 DS232158 EDS36307.1 GDIQ01113524 JAL38202.1 CM000361 CM002910 EDX05738.1 KMY91348.1 CH902620 EDV31872.1 CH964214 EDW80396.1 CH916372 EDV98950.1 CH480833 EDW46461.1 CH954179 EDV54435.2 CH477692 EAT37219.1 AXCM01000826 AE014134 BT150324 AAF53994.3 AGW25606.1 BT011186 AAR88547.1 JARO02004610 KPP68208.1 JRES01000198 KNC33358.1 LRGB01001005 KZS14195.1 AXCN02000752 AP017524 BBB06877.1 CM000158 EDW89934.1
KQ459595 KPI96012.1 RSAL01000051 RVE50258.1 GEYN01000007 JAV02122.1 KQ460323 KPJ15954.1 KQ971345 EFA05437.1 KQ434912 KZC11396.1 MH366154 QBB01963.1 KQ435760 KOX75639.1 NEVH01017446 PNF24469.1 KK853412 KDR07781.1 GL887844 EGI69814.1 IAAA01004539 LAA03364.1 GEZM01057134 JAV72434.1 IAAA01004538 LAA03361.1 KQ980487 KYN15913.1 KQ982409 KYQ56732.1 QCYY01002845 ROT67137.1 KQ982014 KYN30626.1 NNAY01000091 OXU31070.1 KQ977606 KYN01418.1 ADTU01022757 ADTU01022758 KQ976745 KYM75482.1 GL436457 EFN71966.1 KQ414670 KOC64382.1 KK107105 EZA59493.1 QOIP01000011 RLU16408.1 GFJQ02007217 JAV99752.1 GL762206 EFZ21820.1 GDIP01123527 JAL80187.1 GEYN01000008 JAV02121.1 GDIQ01112119 JAL39607.1 AJVK01003753 AJVK01003754 CH933807 KRG03008.1 EDW11968.2 AJWK01015367 NCKU01000824 RWS14003.1 IACF01001021 LAB66751.1 JH432102 GECZ01024196 JAS45573.1 GFXV01002797 MBW14602.1 GDHF01001500 JAI50814.1 GBYB01008111 GBYB01008114 JAG77878.1 JAG77881.1 GAMC01010719 JAB95836.1 OUUW01000010 SPP85985.1 ABLF02036745 ABLF02036754 ABLF02058045 ABLF02059108 CH940654 EDW57959.1 CP012523 ALC39251.1 CR926185 CR926221 BC082822 GL732606 EFX71787.1 GDIP01043744 JAM59971.1 CH479189 EDW25564.1 NCKV01000190 RWS31346.1 BC073490 CH379061 EAL33158.1 CM004472 OCT85802.1 GCES01015798 JAR70525.1 AERX01016021 AERX01016022 DS232158 EDS36307.1 GDIQ01113524 JAL38202.1 CM000361 CM002910 EDX05738.1 KMY91348.1 CH902620 EDV31872.1 CH964214 EDW80396.1 CH916372 EDV98950.1 CH480833 EDW46461.1 CH954179 EDV54435.2 CH477692 EAT37219.1 AXCM01000826 AE014134 BT150324 AAF53994.3 AGW25606.1 BT011186 AAR88547.1 JARO02004610 KPP68208.1 JRES01000198 KNC33358.1 LRGB01001005 KZS14195.1 AXCN02000752 AP017524 BBB06877.1 CM000158 EDW89934.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000283053
UP000053240
UP000007266
+ More
UP000076502 UP000053105 UP000235965 UP000027135 UP000007755 UP000005203 UP000078492 UP000075809 UP000283509 UP000078541 UP000215335 UP000078542 UP000005205 UP000002358 UP000078540 UP000000311 UP000053825 UP000053097 UP000279307 UP000092462 UP000009192 UP000092461 UP000285301 UP000268350 UP000007819 UP000008792 UP000092553 UP000257160 UP000008143 UP000265080 UP000000305 UP000008744 UP000288716 UP000265040 UP000001819 UP000085678 UP000186698 UP000265000 UP000261400 UP000257200 UP000261420 UP000242638 UP000005207 UP000265160 UP000002320 UP000000304 UP000261360 UP000261460 UP000007801 UP000007798 UP000192221 UP000001070 UP000001292 UP000008711 UP000264840 UP000008820 UP000075883 UP000000803 UP000261640 UP000075920 UP000034805 UP000037069 UP000264820 UP000087266 UP000076858 UP000192224 UP000075900 UP000075886 UP000002282
UP000076502 UP000053105 UP000235965 UP000027135 UP000007755 UP000005203 UP000078492 UP000075809 UP000283509 UP000078541 UP000215335 UP000078542 UP000005205 UP000002358 UP000078540 UP000000311 UP000053825 UP000053097 UP000279307 UP000092462 UP000009192 UP000092461 UP000285301 UP000268350 UP000007819 UP000008792 UP000092553 UP000257160 UP000008143 UP000265080 UP000000305 UP000008744 UP000288716 UP000265040 UP000001819 UP000085678 UP000186698 UP000265000 UP000261400 UP000257200 UP000261420 UP000242638 UP000005207 UP000265160 UP000002320 UP000000304 UP000261360 UP000261460 UP000007801 UP000007798 UP000192221 UP000001070 UP000001292 UP000008711 UP000264840 UP000008820 UP000075883 UP000000803 UP000261640 UP000075920 UP000034805 UP000037069 UP000264820 UP000087266 UP000076858 UP000192224 UP000075900 UP000075886 UP000002282
Interpro
IPR009081
PP-bd_ACP
+ More
IPR016161 Ald_DH/histidinol_DH
IPR037022 Formyl_trans_C_sf
IPR036736 ACP-like_sf
IPR016160 Ald_DH_CS_CYS
IPR016163 Ald_DH_C
IPR036477 Formyl_transf_N_sf
IPR002376 Formyl_transf_N
IPR016162 Ald_DH_N
IPR029510 Ald_DH_CS_GLU
IPR005793 Formyl_trans_C
IPR015590 Aldehyde_DH_dom
IPR011407 10_FTHF_DH
IPR011034 Formyl_transferase-like_C_sf
IPR001555 GART_AS
IPR006162 Ppantetheine_attach_site
IPR020806 PKS_PP-bd
IPR016161 Ald_DH/histidinol_DH
IPR037022 Formyl_trans_C_sf
IPR036736 ACP-like_sf
IPR016160 Ald_DH_CS_CYS
IPR016163 Ald_DH_C
IPR036477 Formyl_transf_N_sf
IPR002376 Formyl_transf_N
IPR016162 Ald_DH_N
IPR029510 Ald_DH_CS_GLU
IPR005793 Formyl_trans_C
IPR015590 Aldehyde_DH_dom
IPR011407 10_FTHF_DH
IPR011034 Formyl_transferase-like_C_sf
IPR001555 GART_AS
IPR006162 Ppantetheine_attach_site
IPR020806 PKS_PP-bd
Gene 3D
ProteinModelPortal
A0A2H1W8W8
A0A2A4J0X0
A0A212FMM3
A0A194PRS4
A0A437BIX8
A0A1L8D6P4
+ More
A0A194RER6 D2A5X9 A0A154PHP2 A0A411G7T9 A0A0M9A4E9 A0A2J7Q7B7 A0A067QH51 F4W7E1 A0A087ZYB7 A0A2L2Y5D2 A0A1Y1LFN3 A0A2L2Y5B9 A0A195DSU2 A0A151X8P0 A0A3R7PD39 A0A151JT46 A0A232FKK8 A0A151IHD6 A0A158NPV3 K7ILV9 A0A195ATU6 E2A3S4 A0A0L7R0K3 A0A026WU32 A0A3L8D917 A0A1Z5KX88 E9IC80 A0A0N8CK90 A0A1L8D6Q1 A0A0P5QMT0 A0A1B0GMU0 A0A0Q9X4L0 B4KEZ9 A0A1B0EY70 A0A3S3PF55 A0A2P2HYA6 T1JDA6 A0A1B6F6T2 A0A2H8TLQ5 A0A0K8WIE2 A0A0C9RMA6 W8BRG1 A0A3B0JV67 X1WIZ3 B4M9T3 A0A0M4EA78 A0A3Q1BVF1 Q63ZT8 A0A3P8TBZ4 E9H8V6 A0A0P5ZGX4 B4GT55 A0A443SUZ4 A0A3Q1HEV9 Q6GNL7 Q29KL6 A0A1S3IEW6 A0A1L8GPP8 A0A3Q2QVF3 A0A3B4Z686 A0A3Q1G9C9 A0A3B4U107 A0A146ZX33 A0A3P9QAE5 I3J595 A0A3P9BES5 B0WX46 A0A0P5QR58 B4Q4E1 A0A3B4XY61 A0A3B4FIR4 B3MMB2 B4N7Q7 A0A1W4VAS3 B4JPX0 B4IFN9 B3NLW9 A0A3Q2WDL4 Q16RZ0 A0A182MKG5 Q9VIC9 A0A3Q3NF04 Q6NNU7 A0A182WHJ5 A0A0P7YKU6 A0A0L0CLW3 A0A3Q2Z4G1 A0A1S3P7F6 A0A164XG39 A0A1W4Y042 A0A182RRH5 A0A182Q3G5 A0A2Z5UMA5 B4P6W5
A0A194RER6 D2A5X9 A0A154PHP2 A0A411G7T9 A0A0M9A4E9 A0A2J7Q7B7 A0A067QH51 F4W7E1 A0A087ZYB7 A0A2L2Y5D2 A0A1Y1LFN3 A0A2L2Y5B9 A0A195DSU2 A0A151X8P0 A0A3R7PD39 A0A151JT46 A0A232FKK8 A0A151IHD6 A0A158NPV3 K7ILV9 A0A195ATU6 E2A3S4 A0A0L7R0K3 A0A026WU32 A0A3L8D917 A0A1Z5KX88 E9IC80 A0A0N8CK90 A0A1L8D6Q1 A0A0P5QMT0 A0A1B0GMU0 A0A0Q9X4L0 B4KEZ9 A0A1B0EY70 A0A3S3PF55 A0A2P2HYA6 T1JDA6 A0A1B6F6T2 A0A2H8TLQ5 A0A0K8WIE2 A0A0C9RMA6 W8BRG1 A0A3B0JV67 X1WIZ3 B4M9T3 A0A0M4EA78 A0A3Q1BVF1 Q63ZT8 A0A3P8TBZ4 E9H8V6 A0A0P5ZGX4 B4GT55 A0A443SUZ4 A0A3Q1HEV9 Q6GNL7 Q29KL6 A0A1S3IEW6 A0A1L8GPP8 A0A3Q2QVF3 A0A3B4Z686 A0A3Q1G9C9 A0A3B4U107 A0A146ZX33 A0A3P9QAE5 I3J595 A0A3P9BES5 B0WX46 A0A0P5QR58 B4Q4E1 A0A3B4XY61 A0A3B4FIR4 B3MMB2 B4N7Q7 A0A1W4VAS3 B4JPX0 B4IFN9 B3NLW9 A0A3Q2WDL4 Q16RZ0 A0A182MKG5 Q9VIC9 A0A3Q3NF04 Q6NNU7 A0A182WHJ5 A0A0P7YKU6 A0A0L0CLW3 A0A3Q2Z4G1 A0A1S3P7F6 A0A164XG39 A0A1W4Y042 A0A182RRH5 A0A182Q3G5 A0A2Z5UMA5 B4P6W5
PDB
4GO2
E-value=0,
Score=1964
Ontologies
GO
Topology
Subcellular location
Cytoplasm
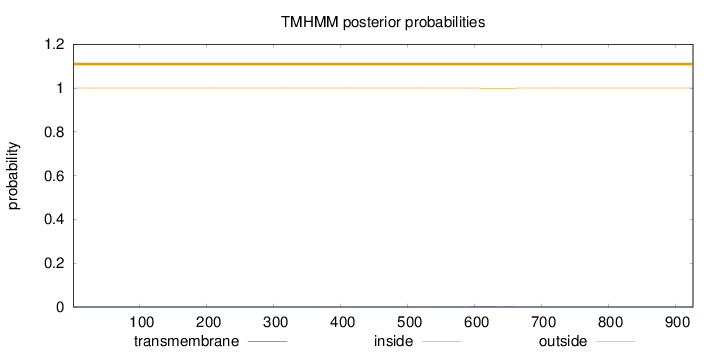
Length:
926
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01671
Exp number, first 60 AAs:
0.00205
Total prob of N-in:
0.00017
outside
1 - 926
Population Genetic Test Statistics
Pi
214.644463
Theta
156.794111
Tajima's D
1.048323
CLR
0.474256
CSRT
0.675066246687666
Interpretation
Uncertain
