Pre Gene Modal
BGIBMGA012995
Annotation
PREDICTED:_L-asparaginase-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 3.471
Sequence
CDS
ATGGCCGAAATAAAACGAGTTCTAGTGCTGTACACTGGAGGTACATTCGGTATGCTGAAAAACGAAAAAGGCGTATTGGTACCGCAAAAAGATATAGAGAGTGTTATTAGGAAACTACCGCAGCTACATGACGATGCATACTGGAAGAACAACCTCGCTGGTACAGATAAGAAGGATTACCTAGCATTACCGGATGGAAAAGACACGGATGTTAAAATTTTGTACCAGATTTACGAATACGATGAACTCAAAGATTCTTCGGATTTCACGATAGACGATTGGATAAAAATGGCGAGAGATATTAAGCGGTTTTACCATGATTACGATGGATTCGTGGTTCTGCACGGCACCGACACAACTGCGTACGGTGGTTCGGTCCTGTCTTTCATGTTGGAAACCGTCGGAAAGACTGTTGTACTCACTGGTGCTCAGGTTCCAATATTTCAACCTCGAAGCGATGGTAACAACAATTTGTTATGCGCTGTGCTGATAGCGGCCACTCAACGCATACCAGAAGTAACAGTGTTCTTTGGAGCAAAACTCTTTAGGGGAACTAGAGTGAAAAAAGTATCGAACACGCGTATTTACGCATTCGACACCCCGAACTTTCCTCCGTTGTTGGAAGCGAAGACCACTTTAGAGGTGGATCCGAAAATGTTGATACATCCTCCAGGATCAGTTCCCAATGAATGCAGGCTACACGACCAGCTATCCAGGAAGGTTTACATTTTGAAGGTGGCGCCTACAATTACACCTGAGTTGATCCGTGGAGTTTGTGAAGGCATGGAAGGGGTAGTTCTAGAGACATATGGAAACGGGAACATACCAATCAAGCGCAAAGAGATCTACAAGGAGATCGAAAGAGCTGTTAAGAACAACGTGTTGGTTGTAAACGTAACTCAATGCATTAACGGTACCGTCCTTGGGAAGGCCATTTACGAAACTGGACTTTTACTCGTGGAATGCGGAGTGGTTCCGGCCTTCGATATGACAGCCGAAGCGGCCTGGGCCAAGTTGTGTTACGTACTGACCAAAACGGAGCTGAGCTATCAACAAAAAGTAGAGTTGATGAAGACAAACATAAGAGGAGAACTGGAAAATCCTAATTCGCGGCACTAA
Protein
MAEIKRVLVLYTGGTFGMLKNEKGVLVPQKDIESVIRKLPQLHDDAYWKNNLAGTDKKDYLALPDGKDTDVKILYQIYEYDELKDSSDFTIDDWIKMARDIKRFYHDYDGFVVLHGTDTTAYGGSVLSFMLETVGKTVVLTGAQVPIFQPRSDGNNNLLCAVLIAATQRIPEVTVFFGAKLFRGTRVKKVSNTRIYAFDTPNFPPLLEAKTTLEVDPKMLIHPPGSVPNECRLHDQLSRKVYILKVAPTITPELIRGVCEGMEGVVLETYGNGNIPIKRKEIYKEIERAVKNNVLVVNVTQCINGTVLGKAIYETGLLLVECGVVPAFDMTAEAAWAKLCYVLTKTELSYQQKVELMKTNIRGELENPNSRH
Summary
Uniprot
H9JTY3
A0A3S2LBW2
A0A2A4K4L8
A0A2H1WGK4
A0A194PRS9
A0A194RES1
+ More
A0A212FMT1 A0A2A4K3H1 A0A194PHI6 A0A0N0PCF4 A0A212ETD0 A0A0P5M800 A0A0N8CBW5 A0A0P6FYU2 A0A0P4YC35 A0A0P4XST9 A0A0N8B0L4 A0A0N7ZJ12 A0A0N8CZL3 A0A0P5RJE8 A0A0P6HI50 A0A0P5TH99 A0A0P5M8G7 A0A0N8CA45 A0A0P5GI12 A0A0P6E3Z9 A0A0P5LTG7 A0A164KJZ7 A0A0L7QTJ1 E9G593 A0A2J7Q619 A0A1V9XKS1 A0A0N0U3X8 A0A226ED56 A0A131XYR0 A0A3L8D7V2 A0A0P5S3V4 A0A0P6BVL9 A0A0P5X8D5 A0A158NTI4 A0A224YS69 A0A2R5LCQ9 A0A0J7L2H7 A0A2H1WRD2 A0A131YM64 F4WG22 A0A154P2Z7 A0A1E1XFI4 B4MAC2 T1P870 A0A1I8MV08 A0A2A3EQ85 A0A1D2NAM8 A0A088AB36 A0A0K2SW72 B3MXE9 E2AI33 A0A224YNE7 A0A1A9W675 A0A0K2SWG8 D6WSZ8 E2C0K9 B4L5C6 A0A2A4JUZ9 A0A0P5BWB9 B4NPB4 A0A1L8E284 A0A131YPN3 A0A0C9QK12 A0A131XFK7 A0A1B0EST4 A0A1B0CBT9 A0A2P2HVG8 L7MKF6 A0A1A9VSV8 A0A034VFD6 A0A034VGZ7 Q9W4N6 A0A1J1HLN0 A0A3S2LU89 A0A1L8E2E3 B3NST3 A0A3B0J8J5 A0A0P5I6U5 B4GW35 L7MFW4 A0A195C797 A0A026WBH7 A0A0N8BV29 Q29G92 A0A1E1XG16 A0A1A9YM98 A0A0L0BKW4 B4I134 A0A1B6EXY1 A0A1B6CX86 A0A1B6FMI7 A0A0M3QZ10 A0A1I8PU47 A0A0A1X7F3
A0A212FMT1 A0A2A4K3H1 A0A194PHI6 A0A0N0PCF4 A0A212ETD0 A0A0P5M800 A0A0N8CBW5 A0A0P6FYU2 A0A0P4YC35 A0A0P4XST9 A0A0N8B0L4 A0A0N7ZJ12 A0A0N8CZL3 A0A0P5RJE8 A0A0P6HI50 A0A0P5TH99 A0A0P5M8G7 A0A0N8CA45 A0A0P5GI12 A0A0P6E3Z9 A0A0P5LTG7 A0A164KJZ7 A0A0L7QTJ1 E9G593 A0A2J7Q619 A0A1V9XKS1 A0A0N0U3X8 A0A226ED56 A0A131XYR0 A0A3L8D7V2 A0A0P5S3V4 A0A0P6BVL9 A0A0P5X8D5 A0A158NTI4 A0A224YS69 A0A2R5LCQ9 A0A0J7L2H7 A0A2H1WRD2 A0A131YM64 F4WG22 A0A154P2Z7 A0A1E1XFI4 B4MAC2 T1P870 A0A1I8MV08 A0A2A3EQ85 A0A1D2NAM8 A0A088AB36 A0A0K2SW72 B3MXE9 E2AI33 A0A224YNE7 A0A1A9W675 A0A0K2SWG8 D6WSZ8 E2C0K9 B4L5C6 A0A2A4JUZ9 A0A0P5BWB9 B4NPB4 A0A1L8E284 A0A131YPN3 A0A0C9QK12 A0A131XFK7 A0A1B0EST4 A0A1B0CBT9 A0A2P2HVG8 L7MKF6 A0A1A9VSV8 A0A034VFD6 A0A034VGZ7 Q9W4N6 A0A1J1HLN0 A0A3S2LU89 A0A1L8E2E3 B3NST3 A0A3B0J8J5 A0A0P5I6U5 B4GW35 L7MFW4 A0A195C797 A0A026WBH7 A0A0N8BV29 Q29G92 A0A1E1XG16 A0A1A9YM98 A0A0L0BKW4 B4I134 A0A1B6EXY1 A0A1B6CX86 A0A1B6FMI7 A0A0M3QZ10 A0A1I8PU47 A0A0A1X7F3
Pubmed
19121390
26354079
22118469
21292972
28327890
29652888
+ More
30249741 21347285 28797301 26830274 21719571 28503490 17994087 18057021 25315136 27289101 20798317 18362917 19820115 28049606 25576852 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 15632085 26108605 25830018
30249741 21347285 28797301 26830274 21719571 28503490 17994087 18057021 25315136 27289101 20798317 18362917 19820115 28049606 25576852 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 15632085 26108605 25830018
EMBL
BABH01003790
RSAL01000051
RVE50260.1
NWSH01000190
PCG78600.1
ODYU01008204
+ More
SOQ51604.1 KQ459595 KPI96017.1 KQ460323 KPJ15959.1 AGBW02007651 OWR55000.1 PCG78599.1 KQ459604 KPI92179.1 KQ460594 KPJ13944.1 AGBW02012595 OWR44746.1 GDIQ01171409 JAK80316.1 GDIQ01094939 JAL56787.1 GDIQ01051720 JAN43017.1 GDIP01229624 JAI93777.1 GDIP01238232 JAI85169.1 GDIQ01224547 JAK27178.1 GDIP01240641 JAI82760.1 GDIP01083343 JAM20372.1 GDIQ01099898 JAL51828.1 GDIQ01018786 JAN75951.1 GDIP01132142 JAL71572.1 GDIQ01182965 JAK68760.1 GDIQ01099899 JAL51827.1 GDIQ01247692 JAK04033.1 GDIQ01068407 JAN26330.1 GDIQ01168518 JAK83207.1 LRGB01003310 KZS03330.1 KQ414741 KOC61963.1 GL732532 EFX85169.1 NEVH01017542 PNF24025.1 MNPL01008915 OQR73982.1 KQ435848 KOX71052.1 LNIX01000004 OXA55533.1 GEFM01004536 GEGO01002753 JAP71260.1 JAR92651.1 QOIP01000012 RLU16364.1 GDIQ01092791 JAL58935.1 GDIP01009029 JAM94686.1 GDIP01076587 JAM27128.1 ADTU01025894 GFPF01009311 MAA20457.1 GGLE01003156 MBY07282.1 LBMM01001132 KMQ96853.1 ODYU01010435 SOQ55536.1 GEDV01008218 JAP80339.1 GL888128 EGI66789.1 KQ434809 KZC06227.1 GFAC01001196 JAT97992.1 CH940655 EDW66181.1 KRF82586.1 KRF82587.1 KRF82588.1 KA644866 AFP59495.1 KZ288194 PBC33850.1 LJIJ01000126 ODN02046.1 HACA01000266 CDW17627.1 CH902630 EDV38414.1 KPU77398.1 GL439621 EFN66941.1 GFPF01006123 MAA17269.1 HACA01000265 CDW17626.1 KQ971352 EFA06327.1 GL451800 EFN78526.1 CH933811 EDW06385.1 NWSH01000611 PCG75313.1 GDIP01178970 JAJ44432.1 CH964291 EDW86354.1 GFDF01001231 JAV12853.1 GEDV01008029 JAP80528.1 GBYB01003879 JAG73646.1 GEFH01003626 JAP64955.1 CCAG010014042 CCAG010014043 AJWK01005824 IACF01000001 LAB65819.1 GACK01000727 JAA64307.1 GAKP01018135 JAC40817.1 GAKP01018134 JAC40818.1 AE014298 AY119242 AAF45911.1 AAM51102.1 CVRI01000010 CRK88968.1 RSAL01000009 RVE53903.1 GFDF01001203 JAV12881.1 CH954180 EDV45763.2 OUUW01000003 SPP78195.1 GDIQ01219250 JAK32475.1 CH479193 EDW26880.1 GACK01002312 JAA62722.1 KQ978205 KYM96510.1 KK107341 EZA52399.1 GDIQ01142027 GDIQ01140122 JAL09699.1 CH379064 EAL32218.2 GFAC01001159 JAT98029.1 JRES01001705 KNC20711.1 CH480819 EDW53215.1 GECZ01026937 JAS42832.1 GEDC01019192 JAS18106.1 GECZ01018390 JAS51379.1 CP012528 ALC48613.1 GBXI01007063 JAD07229.1
SOQ51604.1 KQ459595 KPI96017.1 KQ460323 KPJ15959.1 AGBW02007651 OWR55000.1 PCG78599.1 KQ459604 KPI92179.1 KQ460594 KPJ13944.1 AGBW02012595 OWR44746.1 GDIQ01171409 JAK80316.1 GDIQ01094939 JAL56787.1 GDIQ01051720 JAN43017.1 GDIP01229624 JAI93777.1 GDIP01238232 JAI85169.1 GDIQ01224547 JAK27178.1 GDIP01240641 JAI82760.1 GDIP01083343 JAM20372.1 GDIQ01099898 JAL51828.1 GDIQ01018786 JAN75951.1 GDIP01132142 JAL71572.1 GDIQ01182965 JAK68760.1 GDIQ01099899 JAL51827.1 GDIQ01247692 JAK04033.1 GDIQ01068407 JAN26330.1 GDIQ01168518 JAK83207.1 LRGB01003310 KZS03330.1 KQ414741 KOC61963.1 GL732532 EFX85169.1 NEVH01017542 PNF24025.1 MNPL01008915 OQR73982.1 KQ435848 KOX71052.1 LNIX01000004 OXA55533.1 GEFM01004536 GEGO01002753 JAP71260.1 JAR92651.1 QOIP01000012 RLU16364.1 GDIQ01092791 JAL58935.1 GDIP01009029 JAM94686.1 GDIP01076587 JAM27128.1 ADTU01025894 GFPF01009311 MAA20457.1 GGLE01003156 MBY07282.1 LBMM01001132 KMQ96853.1 ODYU01010435 SOQ55536.1 GEDV01008218 JAP80339.1 GL888128 EGI66789.1 KQ434809 KZC06227.1 GFAC01001196 JAT97992.1 CH940655 EDW66181.1 KRF82586.1 KRF82587.1 KRF82588.1 KA644866 AFP59495.1 KZ288194 PBC33850.1 LJIJ01000126 ODN02046.1 HACA01000266 CDW17627.1 CH902630 EDV38414.1 KPU77398.1 GL439621 EFN66941.1 GFPF01006123 MAA17269.1 HACA01000265 CDW17626.1 KQ971352 EFA06327.1 GL451800 EFN78526.1 CH933811 EDW06385.1 NWSH01000611 PCG75313.1 GDIP01178970 JAJ44432.1 CH964291 EDW86354.1 GFDF01001231 JAV12853.1 GEDV01008029 JAP80528.1 GBYB01003879 JAG73646.1 GEFH01003626 JAP64955.1 CCAG010014042 CCAG010014043 AJWK01005824 IACF01000001 LAB65819.1 GACK01000727 JAA64307.1 GAKP01018135 JAC40817.1 GAKP01018134 JAC40818.1 AE014298 AY119242 AAF45911.1 AAM51102.1 CVRI01000010 CRK88968.1 RSAL01000009 RVE53903.1 GFDF01001203 JAV12881.1 CH954180 EDV45763.2 OUUW01000003 SPP78195.1 GDIQ01219250 JAK32475.1 CH479193 EDW26880.1 GACK01002312 JAA62722.1 KQ978205 KYM96510.1 KK107341 EZA52399.1 GDIQ01142027 GDIQ01140122 JAL09699.1 CH379064 EAL32218.2 GFAC01001159 JAT98029.1 JRES01001705 KNC20711.1 CH480819 EDW53215.1 GECZ01026937 JAS42832.1 GEDC01019192 JAS18106.1 GECZ01018390 JAS51379.1 CP012528 ALC48613.1 GBXI01007063 JAD07229.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000076858 UP000053825 UP000000305 UP000235965 UP000192247 UP000053105 UP000198287 UP000279307 UP000005205 UP000036403 UP000007755 UP000076502 UP000008792 UP000095301 UP000242457 UP000094527 UP000005203 UP000007801 UP000000311 UP000091820 UP000007266 UP000008237 UP000009192 UP000007798 UP000092444 UP000092461 UP000078200 UP000000803 UP000183832 UP000008711 UP000268350 UP000008744 UP000078542 UP000053097 UP000001819 UP000092443 UP000037069 UP000001292 UP000092553 UP000095300
UP000076858 UP000053825 UP000000305 UP000235965 UP000192247 UP000053105 UP000198287 UP000279307 UP000005205 UP000036403 UP000007755 UP000076502 UP000008792 UP000095301 UP000242457 UP000094527 UP000005203 UP000007801 UP000000311 UP000091820 UP000007266 UP000008237 UP000009192 UP000007798 UP000092444 UP000092461 UP000078200 UP000000803 UP000183832 UP000008711 UP000268350 UP000008744 UP000078542 UP000053097 UP000001819 UP000092443 UP000037069 UP000001292 UP000092553 UP000095300
Interpro
IPR027474
L-asparaginase_N
+ More
IPR027473 L-asparaginase_C
IPR040919 Asparaginase_C
IPR041725 L-asparaginase_I
IPR036152 Asp/glu_Ase-like_sf
IPR037152 L-asparaginase_N_sf
IPR006034 Asparaginase/glutaminase-like
IPR027475 Asparaginase/glutaminase_AS2
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR002110 Ankyrin_rpt
IPR006033 AsnA_fam
IPR020827 Asparaginase/glutaminase_AS1
IPR027473 L-asparaginase_C
IPR040919 Asparaginase_C
IPR041725 L-asparaginase_I
IPR036152 Asp/glu_Ase-like_sf
IPR037152 L-asparaginase_N_sf
IPR006034 Asparaginase/glutaminase-like
IPR027475 Asparaginase/glutaminase_AS2
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR002110 Ankyrin_rpt
IPR006033 AsnA_fam
IPR020827 Asparaginase/glutaminase_AS1
Gene 3D
ProteinModelPortal
H9JTY3
A0A3S2LBW2
A0A2A4K4L8
A0A2H1WGK4
A0A194PRS9
A0A194RES1
+ More
A0A212FMT1 A0A2A4K3H1 A0A194PHI6 A0A0N0PCF4 A0A212ETD0 A0A0P5M800 A0A0N8CBW5 A0A0P6FYU2 A0A0P4YC35 A0A0P4XST9 A0A0N8B0L4 A0A0N7ZJ12 A0A0N8CZL3 A0A0P5RJE8 A0A0P6HI50 A0A0P5TH99 A0A0P5M8G7 A0A0N8CA45 A0A0P5GI12 A0A0P6E3Z9 A0A0P5LTG7 A0A164KJZ7 A0A0L7QTJ1 E9G593 A0A2J7Q619 A0A1V9XKS1 A0A0N0U3X8 A0A226ED56 A0A131XYR0 A0A3L8D7V2 A0A0P5S3V4 A0A0P6BVL9 A0A0P5X8D5 A0A158NTI4 A0A224YS69 A0A2R5LCQ9 A0A0J7L2H7 A0A2H1WRD2 A0A131YM64 F4WG22 A0A154P2Z7 A0A1E1XFI4 B4MAC2 T1P870 A0A1I8MV08 A0A2A3EQ85 A0A1D2NAM8 A0A088AB36 A0A0K2SW72 B3MXE9 E2AI33 A0A224YNE7 A0A1A9W675 A0A0K2SWG8 D6WSZ8 E2C0K9 B4L5C6 A0A2A4JUZ9 A0A0P5BWB9 B4NPB4 A0A1L8E284 A0A131YPN3 A0A0C9QK12 A0A131XFK7 A0A1B0EST4 A0A1B0CBT9 A0A2P2HVG8 L7MKF6 A0A1A9VSV8 A0A034VFD6 A0A034VGZ7 Q9W4N6 A0A1J1HLN0 A0A3S2LU89 A0A1L8E2E3 B3NST3 A0A3B0J8J5 A0A0P5I6U5 B4GW35 L7MFW4 A0A195C797 A0A026WBH7 A0A0N8BV29 Q29G92 A0A1E1XG16 A0A1A9YM98 A0A0L0BKW4 B4I134 A0A1B6EXY1 A0A1B6CX86 A0A1B6FMI7 A0A0M3QZ10 A0A1I8PU47 A0A0A1X7F3
A0A212FMT1 A0A2A4K3H1 A0A194PHI6 A0A0N0PCF4 A0A212ETD0 A0A0P5M800 A0A0N8CBW5 A0A0P6FYU2 A0A0P4YC35 A0A0P4XST9 A0A0N8B0L4 A0A0N7ZJ12 A0A0N8CZL3 A0A0P5RJE8 A0A0P6HI50 A0A0P5TH99 A0A0P5M8G7 A0A0N8CA45 A0A0P5GI12 A0A0P6E3Z9 A0A0P5LTG7 A0A164KJZ7 A0A0L7QTJ1 E9G593 A0A2J7Q619 A0A1V9XKS1 A0A0N0U3X8 A0A226ED56 A0A131XYR0 A0A3L8D7V2 A0A0P5S3V4 A0A0P6BVL9 A0A0P5X8D5 A0A158NTI4 A0A224YS69 A0A2R5LCQ9 A0A0J7L2H7 A0A2H1WRD2 A0A131YM64 F4WG22 A0A154P2Z7 A0A1E1XFI4 B4MAC2 T1P870 A0A1I8MV08 A0A2A3EQ85 A0A1D2NAM8 A0A088AB36 A0A0K2SW72 B3MXE9 E2AI33 A0A224YNE7 A0A1A9W675 A0A0K2SWG8 D6WSZ8 E2C0K9 B4L5C6 A0A2A4JUZ9 A0A0P5BWB9 B4NPB4 A0A1L8E284 A0A131YPN3 A0A0C9QK12 A0A131XFK7 A0A1B0EST4 A0A1B0CBT9 A0A2P2HVG8 L7MKF6 A0A1A9VSV8 A0A034VFD6 A0A034VGZ7 Q9W4N6 A0A1J1HLN0 A0A3S2LU89 A0A1L8E2E3 B3NST3 A0A3B0J8J5 A0A0P5I6U5 B4GW35 L7MFW4 A0A195C797 A0A026WBH7 A0A0N8BV29 Q29G92 A0A1E1XG16 A0A1A9YM98 A0A0L0BKW4 B4I134 A0A1B6EXY1 A0A1B6CX86 A0A1B6FMI7 A0A0M3QZ10 A0A1I8PU47 A0A0A1X7F3
PDB
4R8L
E-value=4.02186e-67,
Score=646
Ontologies
GO
Topology
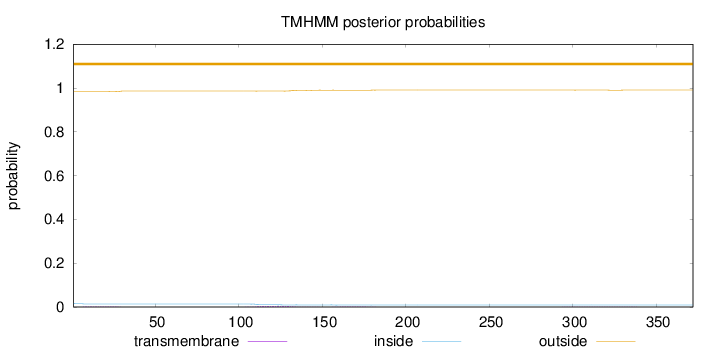
Length:
372
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.19094
Exp number, first 60 AAs:
0.04012
Total prob of N-in:
0.01566
outside
1 - 372
Population Genetic Test Statistics
Pi
274.349169
Theta
154.080373
Tajima's D
2.404469
CLR
25.4798
CSRT
0.937253137343133
Interpretation
Uncertain
