Pre Gene Modal
BGIBMGA012996
Annotation
Glucose_dehydrogenase_[acceptor]?_partial_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 1.19 Mitochondrial Reliability : 1.124
Sequence
CDS
ATGAAAGGGGGACGCTGCAACTGGCCTCGTGGTAAAGTCCTGGGTGGCAGTTCCGTTTTGAACTACATGCTCTATCTTCGAGGTAATAGAAGAGATTATGACACATGGGAGGCGCTGGGCAACAAAGGATGGGGTTATAAGGACGTGCTTCATTACTTCAAGAAATCAGAAGATAACAAGAATCCGTATTTAGCTCAAACACCTTATCACAGTACCGGTGGATACCTTACGGTTGCAGAAGCCCCTTACCAAACGCCACTAGTGTCGAGTTTCATTGAAGGTGGATTAGAATTAGGGTACATGAACCGAGACATAAACGGCGAAAACCAGACAGGCTTTATGGTAGCCCAAGGCACTGTCAGACGAGGCAGCCGTTGTTCTACTTCAAAGGCTTTTCTGAGATCCGCTAAAGATCGCCAAAACTTACACATATCTATGCATTCTCACGTTACTAAAGTTCTAATAGATCCACGCACGAAAATAGCATTTGGTGTTGAATTTATACGCGATAAGATGGTACACCGCATCAGAGCACGAAAAGAGGTAATACTTTCCGCTGGTACCATCAACTCAGCTCAAATTTTAATGTTATCTGGCATCGGTCCAAGTGAAGAACTAGCAAAGCATCGGATACCTGTTATCCAGAATCTAAAGGTTGGCAGTAATTTGCAAGATCACATTGGTTTAGGCGGTTTAGCCTTCATGGTAAACAAAGAAATTTCAATTGTCGAACACCGCTTACATACAGTGAGCACTTTGATGGAGTACGCAGTGCTCGGTGAAGGTCCCTTGACAATTATGGGCGGAGTCGAGGGTCTAGCATTCGTGAATACGAAATACGCAAATGCTTCAGACGATTTTCCTGACATTGAATTCCACTTCATATCCGGTTCAACAAATTCCGACGGCGGTGAACAAATACGTAAAGCACACGGGCTGATGGAGAATTTTTATGATGCCGTTTTTAGACCGATTAACAATATGGACGTTTGGAGTATCATTCCGATGTTGCTACGTCCAAGAAGCAAAGGTTTTATACGGTTACGCAGCTCAAACCCATACGATTATCCATACATTTATCCAAATTATTTGATGGACGAAATGGATGTTAAGACGCTTGTTGAAGGCGTTAAAATCGCTATAGCATTGTCTAGGACACAAGCTTTCCAACGACATGGTTCCGTTTTGAATAAACACGTCTTTCCTGCGTGTGTTAGTATTAAGAGGTACACAGATGCGTATTGGGAATGCATGATAAGGCAGTACACGTGCACAATTTACCATCCAGTCGGCACTGCGAAAATGGGACCCTACTGGGATCCAGATGCAGTCGTTGACCCCGAACTGAGAGTGTATGGAATCAAAGGCCTTAGGGTAATCGATGGTAGCATAATGCCTAATTTGGTTAGTGGAAATACTAATGCACCAATTATTATGATTGGCGAAAAAGGCAGTGATATGATCAAAGAGTTCTGCCCAGTAAAAAGTATAGTTTCGGGCACCAGAATTTTGTTTGGTTTACTCCCAGGTTTGGGTGTCCTGATCGTGATCCGTATGGCTATACATTTGTACAGACCCGATATTGAAGATTCAGAGCATCGGGTCAGGGATCACCCGATCGAAGACCTGAACGATTGCTACGATTTTATCATCGTCGGTGGTGGTTCTGCTGGAGCGGTTTTAGCGAATCGTCTGACAGAAAACCCAGCATGGACAGTACTGTTATTAGAAGCTGGACCCGATGAGAATGTGCTTTCCGAAGTGCCCGTGCTGTTTCCGATATTACAAACTTCATTTATAGACTGGCAGTTTGTGACCGAGCCCAGTGATGATTATTGTCTCAGTATGATTGACAAAAGGTGTAAGTGGCCACGCGGTAAAGTTCTCGGTGGATCTAGTGTTCTAAATGCAATGTTATATATTAGAGGCAATAAACGTGATTACGATAACTGGAAAAGAATGGGAAACGCCGGTTGGAGTTACAACGATGTATTCCCCTACTTTTTGAAATCTGAAGATATCAGAATTGCAGAGCATCAAAATGATTCTTATCATGCAACTGGAGGTCCGTTAACCATAGAATATTTTAGATACGAGCATCCAATCACAAACAAGATTTTAGAAGCAGGCGCACAACTTGGTTACGTAGTAAGGGATGTAAACGGTGAATTTCAAACAGGATTCACTCGCTCTCAGGCAACCGTGCGAGATGGTCTTCGTTGCAGCACAGCAAAGGCATTTCTAAGACCTATATCTAAAAGAAAGAATCTTCACATCAGTATGAACTCTTTAGTAGAAAAAGTGCTGATTGATGAGTACCGAACAGTTTATGGGGTAAAGTTTTCAAAGCATGGTCATAAAAAAATCGTGAAAGCCAGCAAGGAAGTAATTCTATCGGCCGGTTCTATTCAGTCGCCGCAATTATTGATGTTATCTGGTATCGGCGATGCTGCTGAATTAAAAGAACTAGGGATATTTCCAATAGCACATCTGCCAGGAGTTGGGAAAAATTTACAAGACCATACTGCTATGGGCGGTCACTCCTTTCTTTTTGAGTACCCTTCTGACAAAGGAGCTCGTTTTTGCTTTGACCTAAATACTTTATTTTCTTTCGGAAATCTTATTGACTTTTGTATTCATAAAAGTGGATTAATGTACAGTATGGTAGAGGCTGAGGCAATGGCTTTTGTAAACACGAAATATCAAAACCCAGCTGAAGATTATCCCGATATACAATTATTTATTGCTCCGACAGCAGATAATATGGACGGCGGTCTTTTTGGGAAACGAGCTAATGGTATCACAGACGAAACATACGCTGAATTATATGAGGATCTATTATACGAATCATCGTTTTCAATCGTTCCACTTTTACTAAGACCAAAAAGCCGAGGCTACATCAAATTGCGGGATAGCAATCCTAATTCACCTCCTCTTATTTATCCAAACTACTATTCCCATCCAGACGATGTGAAGATTATGGTGGAAGGAGCCCGTATAGCACAAGCATTGGTAGAACAACCAGCACTAAAAGAAATGAATGCTCGACCCAACCCACATCGTAACCCTGGTTGCGAGGAGCATGAGTTGATGTCTTTCGAACACTTGGAATGTCAAGCCCGGCATCATACTCTGACCATCTATCATCCGGTAGGAACTTGCGCTATGGGCCCGCCCGACAATCCAGAGGCTGTTGTGGATGCAAGATTACGGGTACGAATGGTATCTTCATTAGATTTGTATGTAAATATTAAAGGTTCTAATTTAAAACCGACCATTGATCTGTTTTAG
Protein
MKGGRCNWPRGKVLGGSSVLNYMLYLRGNRRDYDTWEALGNKGWGYKDVLHYFKKSEDNKNPYLAQTPYHSTGGYLTVAEAPYQTPLVSSFIEGGLELGYMNRDINGENQTGFMVAQGTVRRGSRCSTSKAFLRSAKDRQNLHISMHSHVTKVLIDPRTKIAFGVEFIRDKMVHRIRARKEVILSAGTINSAQILMLSGIGPSEELAKHRIPVIQNLKVGSNLQDHIGLGGLAFMVNKEISIVEHRLHTVSTLMEYAVLGEGPLTIMGGVEGLAFVNTKYANASDDFPDIEFHFISGSTNSDGGEQIRKAHGLMENFYDAVFRPINNMDVWSIIPMLLRPRSKGFIRLRSSNPYDYPYIYPNYLMDEMDVKTLVEGVKIAIALSRTQAFQRHGSVLNKHVFPACVSIKRYTDAYWECMIRQYTCTIYHPVGTAKMGPYWDPDAVVDPELRVYGIKGLRVIDGSIMPNLVSGNTNAPIIMIGEKGSDMIKEFCPVKSIVSGTRILFGLLPGLGVLIVIRMAIHLYRPDIEDSEHRVRDHPIEDLNDCYDFIIVGGGSAGAVLANRLTENPAWTVLLLEAGPDENVLSEVPVLFPILQTSFIDWQFVTEPSDDYCLSMIDKRCKWPRGKVLGGSSVLNAMLYIRGNKRDYDNWKRMGNAGWSYNDVFPYFLKSEDIRIAEHQNDSYHATGGPLTIEYFRYEHPITNKILEAGAQLGYVVRDVNGEFQTGFTRSQATVRDGLRCSTAKAFLRPISKRKNLHISMNSLVEKVLIDEYRTVYGVKFSKHGHKKIVKASKEVILSAGSIQSPQLLMLSGIGDAAELKELGIFPIAHLPGVGKNLQDHTAMGGHSFLFEYPSDKGARFCFDLNTLFSFGNLIDFCIHKSGLMYSMVEAEAMAFVNTKYQNPAEDYPDIQLFIAPTADNMDGGLFGKRANGITDETYAELYEDLLYESSFSIVPLLLRPKSRGYIKLRDSNPNSPPLIYPNYYSHPDDVKIMVEGARIAQALVEQPALKEMNARPNPHRNPGCEEHELMSFEHLECQARHHTLTIYHPVGTCAMGPPDNPEAVVDARLRVRMVSSLDLYVNIKGSNLKPTIDLF
Summary
Similarity
Belongs to the GMC oxidoreductase family.
Uniprot
Pubmed
EMBL
BABH01003796
BABH01003797
JTDY01010401
KOB58222.1
NNAY01000267
OXU29612.1
+ More
KB632330 ERL92579.1 APGK01055694 APGK01055695 APGK01055696 APGK01055697 APGK01055698 APGK01055699 APGK01055700 APGK01055701 APGK01055702 APGK01055703 KB741269 ENN71447.1 KQ978473 KYM93715.1 CVRI01000064 CRL05333.1 PYGN01000027 PSN57124.1 AGBW02007651 OWR55009.1 KK852802 KDR16285.1 KK107261 EZA53943.1 GL448571 EFN84155.1 OXU29610.1 KQ971348 EFA05534.2 OWR55004.1
KB632330 ERL92579.1 APGK01055694 APGK01055695 APGK01055696 APGK01055697 APGK01055698 APGK01055699 APGK01055700 APGK01055701 APGK01055702 APGK01055703 KB741269 ENN71447.1 KQ978473 KYM93715.1 CVRI01000064 CRL05333.1 PYGN01000027 PSN57124.1 AGBW02007651 OWR55009.1 KK852802 KDR16285.1 KK107261 EZA53943.1 GL448571 EFN84155.1 OXU29610.1 KQ971348 EFA05534.2 OWR55004.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF51905
SSF51905
Gene 3D
ProteinModelPortal
PDB
5OC1
E-value=1.19353e-51,
Score=518
Ontologies
GO
Topology
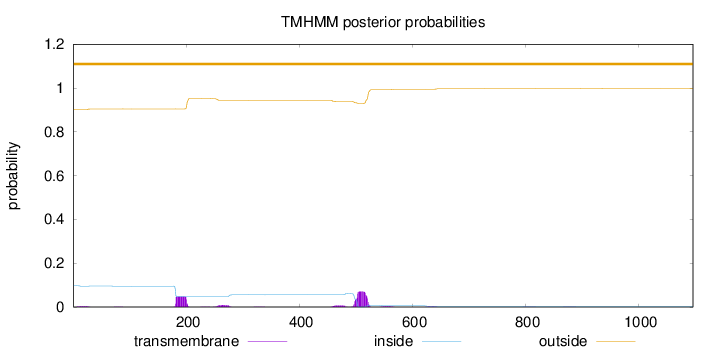
Length:
1096
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.94585
Exp number, first 60 AAs:
0.06175
Total prob of N-in:
0.09834
outside
1 - 1096
Population Genetic Test Statistics
Pi
187.612743
Theta
169.933934
Tajima's D
0.083812
CLR
21.68674
CSRT
0.394130293485326
Interpretation
Uncertain
