Gene
KWMTBOMO09586
Pre Gene Modal
BGIBMGA012999
Annotation
PREDICTED:_glucose_dehydrogenase_[FAD?_quinone]-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.447
Sequence
CDS
ATGTCGGCGTGTGATCCGACGTTCACTTCGACGGTGATAGGCAGTTACGGAGACGTGGTTGGGACCACGTTCGCTCAAGCACTCACTAGTTTCCTCGCATTACGGAGACGTGAGGATGCTATTAACTCTGTTGTCGATGGTAGTAGCTATGATTTTGTGGTGGTCGGAGCAGGATCAGCCGGTTCAGTCTTGGCAAACCGTTTAAGTGAAGTCCCAGAATGGAAAGTATTACTAGTGGAAGCTGGAGGAAACCCTACCGTGAACACTGAGATACCACAAGTCTTTTATAACAACTTGGGCAGTAAAGAAGACTGGGGTTACCGCCCAGAACCGCAATCCGGCGCTTGTCTCAGTTATGAAACTAAAACATGTGCCTGGCCTAGTGGGAAGACACTCGGTGGCTCCGCAAGTATTAATGCTATGTTCTATGTCAGAGGAAACCACATAGACTTCGATGGATGGGCAGAAGACGGCAACAACGGTTGGAGTTACGAAGATGTTCTTCCTTACTTTAAAAAAAGTGAAAATTTTGTAGCAGATATCTCAGAAAACGATAGATATCATGGCACTAAAGGTGAATTATATACCACAAAAGATAAAAACTTAAAGCCTTTTGAAGAGGTGGTTCTTAAAGCATACCCAGAGTTAGGAGTACAAAATTTAGACGATATAAACGGAGAAAATCAAATGGGCGTAACACAAAGCCAAATTACTGTTTACAACAAAACAAGAATGAGTACCGCTAGAGCGTTCCTTTCGCCAATCCGGAATAGAAACAATTTGCATGTTATCAAAAATACACTGGCTACGAAAATTATATTTAAAGAGAACAGTAATGCCGTTAGTGGAATTATGCTAAACAACAACGGCAAAGAATTTATGGTTAATGTGAACAAAGAGTTGATATTGTCAGCGGGAGCAATTAATTCACCGCACCTTTTAATGTTGTCCGGTATTGGACCTCAAGAACACCTACTTGAACAAAATATTACTGTCAAAATGGATTTAGCGGTTGGAGAAAATCTACAAGATCATGTCTTCGTGCCATTATTGTATACAGCCCCTGCCAAAGAATCTGACGATATTACTATGAGCTTAGTCGTAAACAACTTTTTCGATTACATGGTTAATAAAGAAGGAATACTTTGTGACACCAGTCCGCATAGAATAATTGCATTCGTCAATACCACTGATTCAACTGGTACGAGTCCTGACTTACAATTTCATCACGTTATTTTTACACCTAAAATTCACATTTTACTAGACTTTTTTGAGAAACACGGACTAAATAATGATATTATACAACAATTCCATGAAATTAACAACAATAAGTTCGCTATGTTAATCTACAATACTTTATTGAAACCAAAATCACGAGGAAAAGTATTACTGCAAAGCAATAACCCATTTGATCGTCCATTGATATACGCCAACTATTTCAATGATAATGAAGACATGAAGACGTTAATAAGGGGTATGAGGAAATTTACTTTGAAATTAGGCGAAACAGACGCCTTCAAACAAGTAGGGTTAGAATTAGAATGGTTTCAAATACCAACCTGTGATAAACTTGATAAAACAAGTGACGAATTTTTGGAGTGCATTTCGAGACACTTAACTTTTTCATTGTATCACCCATCGGGCACCGTAAAGATGGGACCAGAAGACGATCCTAGCGCAGTAGTGACTCCTGAGTTAAAAGTTCGAAATATTGAAAATTTGAGAGTAATTGACGCTAGTGTTATGCCTTCCATTGTAAGAGGTAATACTAATGCAGCGACAATCATGATTGGAGAAAAAGGTGCAGAACTAATCAAACAATCCCACATAAAAACAACAGAATAA
Protein
MSACDPTFTSTVIGSYGDVVGTTFAQALTSFLALRRREDAINSVVDGSSYDFVVVGAGSAGSVLANRLSEVPEWKVLLVEAGGNPTVNTEIPQVFYNNLGSKEDWGYRPEPQSGACLSYETKTCAWPSGKTLGGSASINAMFYVRGNHIDFDGWAEDGNNGWSYEDVLPYFKKSENFVADISENDRYHGTKGELYTTKDKNLKPFEEVVLKAYPELGVQNLDDINGENQMGVTQSQITVYNKTRMSTARAFLSPIRNRNNLHVIKNTLATKIIFKENSNAVSGIMLNNNGKEFMVNVNKELILSAGAINSPHLLMLSGIGPQEHLLEQNITVKMDLAVGENLQDHVFVPLLYTAPAKESDDITMSLVVNNFFDYMVNKEGILCDTSPHRIIAFVNTTDSTGTSPDLQFHHVIFTPKIHILLDFFEKHGLNNDIIQQFHEINNNKFAMLIYNTLLKPKSRGKVLLQSNNPFDRPLIYANYFNDNEDMKTLIRGMRKFTLKLGETDAFKQVGLELEWFQIPTCDKLDKTSDEFLECISRHLTFSLYHPSGTVKMGPEDDPSAVVTPELKVRNIENLRVIDASVMPSIVRGNTNAATIMIGEKGAELIKQSHIKTTE
Summary
Cofactor
FAD
Similarity
Belongs to the GMC oxidoreductase family.
Uniprot
H9JTY7
H9JTY6
H9JTY8
H9JTY5
I6YR69
A0A2A4K4A7
+ More
A0A2H1VTB0 A0A2A4K4K5 A0A3S2P1X7 A0A194PSS7 A0A194RDP5 A0A194PRT5 A0A0L7KNL7 I4DNK7 A0A194R0E8 A0A1B0GIG3 A0A1B6EWM9 A0A1B6EYD4 A0A0A9YGE8 A0A1B6HJ69 A0A1B6K5Y4 A0A2J7PXM4 A0A2J7PXQ0 A0A1B6KJY4 A0A1B6LHN9 A0A151IVV0 A0A1B6G282 A0A1B6JVY6 A0A2J7RBI7 A0A1L8E0M0 A0A1L8E037 A0A0J7K353 A0A2P8ZHS7 A0A195BBJ1 A0A1B6DKE4 E2BJJ0 A0A1L8E0E7 A0A067R8P7 A0A158NUQ9 A0A1L8DZX6 Q17DW2 A0A0P5PUH8 A0A151K093 A0A2M4ALF8 A0A0P5TWX8 A0A0P5UXE2 A0A0P5C9U0 A0A151K0E9 A0A336MWJ3 A0A151WNR9 A0A1B0D7C5 A0A026WCZ8 A0A0P5UXN7 A0A2M4AF63 E0VUB2 A0A164QA14 E2ASE9 A0A023EVK5 A0A3L8D7W0 A0A182FZT6 A0A151WMQ2 A0A0N8CY00 A0A2J7PXM0 A0A088AJU7 A0A0N7ZRH0 A0A0P4YIW2 A0A0P5WPU7 F4WR74 A0A0P5UVG6 A0A0P5JJB6 A0A0P5HR00 A0A151J323 A0A0P5Q3U3 F5HJA8 A0A2M4AFT0 A0A0P5MEE8 E1ZVM3 A0A0P5GLG5 A0A0P5UVJ4 A0A0P4ZXT5 A0A0P5LXN9 A0A0P5KVV5 A0A0P5QPX0 A0A0P5C9R7 A0A0P5FKY8 A0A0P5RT58 A0A1B6HLV3 A0A2M4AHF2 A0A0P4ZIS6 A0A0A1WEA8 A0A0P5C863 A0A0P6E0K9 A0A0J7KLX4 A0A0P6D8A7 A0A453YNJ2 A0A0P5IKE5 B4JKZ4 A0A0N8DJ56 A0A0P5M7L3 A0A0N8C666
A0A2H1VTB0 A0A2A4K4K5 A0A3S2P1X7 A0A194PSS7 A0A194RDP5 A0A194PRT5 A0A0L7KNL7 I4DNK7 A0A194R0E8 A0A1B0GIG3 A0A1B6EWM9 A0A1B6EYD4 A0A0A9YGE8 A0A1B6HJ69 A0A1B6K5Y4 A0A2J7PXM4 A0A2J7PXQ0 A0A1B6KJY4 A0A1B6LHN9 A0A151IVV0 A0A1B6G282 A0A1B6JVY6 A0A2J7RBI7 A0A1L8E0M0 A0A1L8E037 A0A0J7K353 A0A2P8ZHS7 A0A195BBJ1 A0A1B6DKE4 E2BJJ0 A0A1L8E0E7 A0A067R8P7 A0A158NUQ9 A0A1L8DZX6 Q17DW2 A0A0P5PUH8 A0A151K093 A0A2M4ALF8 A0A0P5TWX8 A0A0P5UXE2 A0A0P5C9U0 A0A151K0E9 A0A336MWJ3 A0A151WNR9 A0A1B0D7C5 A0A026WCZ8 A0A0P5UXN7 A0A2M4AF63 E0VUB2 A0A164QA14 E2ASE9 A0A023EVK5 A0A3L8D7W0 A0A182FZT6 A0A151WMQ2 A0A0N8CY00 A0A2J7PXM0 A0A088AJU7 A0A0N7ZRH0 A0A0P4YIW2 A0A0P5WPU7 F4WR74 A0A0P5UVG6 A0A0P5JJB6 A0A0P5HR00 A0A151J323 A0A0P5Q3U3 F5HJA8 A0A2M4AFT0 A0A0P5MEE8 E1ZVM3 A0A0P5GLG5 A0A0P5UVJ4 A0A0P4ZXT5 A0A0P5LXN9 A0A0P5KVV5 A0A0P5QPX0 A0A0P5C9R7 A0A0P5FKY8 A0A0P5RT58 A0A1B6HLV3 A0A2M4AHF2 A0A0P4ZIS6 A0A0A1WEA8 A0A0P5C863 A0A0P6E0K9 A0A0J7KLX4 A0A0P6D8A7 A0A453YNJ2 A0A0P5IKE5 B4JKZ4 A0A0N8DJ56 A0A0P5M7L3 A0A0N8C666
Pubmed
EMBL
BABH01003798
BABH01003799
BABH01003797
JQ965926
AFN71166.1
NWSH01000190
+ More
PCG78593.1 ODYU01004023 SOQ43474.1 PCG78592.1 RSAL01000051 RVE50264.1 KQ459595 KPI96023.1 KQ460323 KPJ15963.1 KPI96022.1 JTDY01008426 KOB64574.1 AK402986 BAM19497.1 KQ460890 KPJ10999.1 AJWK01014080 GECZ01027417 JAS42352.1 GECZ01026787 JAS42982.1 GBHO01012330 GBRD01008695 GBRD01008694 JAG31274.1 JAG57126.1 GECU01033045 JAS74661.1 GECU01001143 JAT06564.1 NEVH01020853 PNF21087.1 PNF21103.1 GEBQ01028513 JAT11464.1 GEBQ01016745 JAT23232.1 KQ980881 KYN11981.1 GECZ01013224 JAS56545.1 GECU01004364 JAT03343.1 NEVH01005904 PNF38181.1 GFDF01002039 JAV12045.1 GFDF01002040 JAV12044.1 LBMM01015561 KMQ84762.1 PYGN01000053 PSN56054.1 KQ976532 KYM81564.1 GEDC01024026 GEDC01011147 JAS13272.1 JAS26151.1 GL448571 EFN84147.1 GFDF01002042 JAV12042.1 KK852811 KDR15959.1 ADTU01026683 GFDF01002041 JAV12043.1 CH477290 EAT44639.1 GDIQ01125583 JAL26143.1 KQ981296 KYN43077.1 GGFK01008304 MBW41625.1 GDIP01122942 GDIQ01046714 JAL80772.1 JAN48023.1 GDIP01107288 JAL96426.1 GDIP01189775 GDIP01189774 GDIP01109548 JAJ33628.1 KYN43080.1 UFQT01002442 SSX33469.1 KQ982905 KYQ49514.1 AJVK01026844 AJVK01026845 AJVK01026846 KK107261 EZA53937.1 GDIP01107286 JAL96428.1 GGFK01006105 MBW39426.1 DS235784 EEB16968.1 LRGB01002451 KZS07567.1 GL442298 EFN63615.1 GAPW01000526 JAC13072.1 QOIP01000012 RLU16172.1 KQ982937 KYQ49100.1 GDIP01087847 JAM15868.1 PNF21089.1 GDIP01219777 JAJ03625.1 GDIP01228120 JAI95281.1 GDIP01083296 JAM20419.1 GL888284 EGI63349.1 GDIP01109547 JAL94167.1 GDIQ01223788 JAK27937.1 GDIQ01223789 GDIQ01223787 JAK27936.1 KQ980314 KYN16692.1 GDIQ01126533 JAL25193.1 AAAB01008844 EGK96369.1 GGFK01006251 MBW39572.1 GDIQ01251280 GDIQ01251279 GDIQ01185200 GDIQ01182102 GDIQ01180798 GDIQ01163719 GDIQ01163718 GDIQ01163715 JAK70927.1 GL434548 EFN74792.1 GDIQ01242702 JAK09023.1 GDIP01108083 JAL95631.1 GDIP01228121 GDIP01219927 GDIP01174575 GDIP01158438 JAJ03475.1 KZS07507.1 GDIQ01251278 GDIQ01188299 GDIQ01185202 GDIQ01185201 GDIQ01180799 GDIQ01163717 GDIQ01163716 GDIQ01094633 JAK88009.1 GDIQ01204286 JAK47439.1 GDIQ01162366 GDIQ01111223 JAL40503.1 GDIP01177549 JAJ45853.1 GDIQ01253882 JAJ97842.1 GDIQ01216300 GDIQ01162367 GDIQ01162365 GDIQ01157523 GDIQ01112823 GDIQ01096486 GDIQ01031667 JAL55240.1 GECU01032039 JAS75667.1 GGFK01006898 MBW40219.1 GDIP01216074 JAJ07328.1 GBXI01017045 GBXI01016813 GBXI01008932 JAC97246.1 JAC97478.1 JAD05360.1 GDIP01174531 JAJ48871.1 GDIQ01248185 GDIQ01241251 GDIQ01220935 GDIQ01196405 GDIQ01177948 GDIQ01174041 GDIQ01142131 GDIQ01116533 GDIQ01115331 GDIQ01109424 GDIQ01073988 GDIQ01069503 GDIQ01069502 GDIQ01069501 GDIQ01067900 GDIQ01059899 GDIQ01057405 JAN25236.1 LBMM01005695 KMQ91287.1 GDIQ01082738 JAN11999.1 GDIQ01212735 JAK38990.1 CH916370 EDW00247.1 GDIP01028599 JAM75116.1 GDIQ01222435 GDIQ01222434 GDIQ01220936 GDIQ01220934 GDIQ01220933 GDIQ01212734 GDIQ01204243 GDIQ01186800 GDIQ01186799 GDIQ01183405 GDIQ01183404 GDIQ01183403 GDIQ01155173 GDIQ01152368 GDIQ01123933 GDIQ01067899 GDIQ01066404 GDIQ01063605 JAK68321.1 GDIQ01110931 JAL40795.1
PCG78593.1 ODYU01004023 SOQ43474.1 PCG78592.1 RSAL01000051 RVE50264.1 KQ459595 KPI96023.1 KQ460323 KPJ15963.1 KPI96022.1 JTDY01008426 KOB64574.1 AK402986 BAM19497.1 KQ460890 KPJ10999.1 AJWK01014080 GECZ01027417 JAS42352.1 GECZ01026787 JAS42982.1 GBHO01012330 GBRD01008695 GBRD01008694 JAG31274.1 JAG57126.1 GECU01033045 JAS74661.1 GECU01001143 JAT06564.1 NEVH01020853 PNF21087.1 PNF21103.1 GEBQ01028513 JAT11464.1 GEBQ01016745 JAT23232.1 KQ980881 KYN11981.1 GECZ01013224 JAS56545.1 GECU01004364 JAT03343.1 NEVH01005904 PNF38181.1 GFDF01002039 JAV12045.1 GFDF01002040 JAV12044.1 LBMM01015561 KMQ84762.1 PYGN01000053 PSN56054.1 KQ976532 KYM81564.1 GEDC01024026 GEDC01011147 JAS13272.1 JAS26151.1 GL448571 EFN84147.1 GFDF01002042 JAV12042.1 KK852811 KDR15959.1 ADTU01026683 GFDF01002041 JAV12043.1 CH477290 EAT44639.1 GDIQ01125583 JAL26143.1 KQ981296 KYN43077.1 GGFK01008304 MBW41625.1 GDIP01122942 GDIQ01046714 JAL80772.1 JAN48023.1 GDIP01107288 JAL96426.1 GDIP01189775 GDIP01189774 GDIP01109548 JAJ33628.1 KYN43080.1 UFQT01002442 SSX33469.1 KQ982905 KYQ49514.1 AJVK01026844 AJVK01026845 AJVK01026846 KK107261 EZA53937.1 GDIP01107286 JAL96428.1 GGFK01006105 MBW39426.1 DS235784 EEB16968.1 LRGB01002451 KZS07567.1 GL442298 EFN63615.1 GAPW01000526 JAC13072.1 QOIP01000012 RLU16172.1 KQ982937 KYQ49100.1 GDIP01087847 JAM15868.1 PNF21089.1 GDIP01219777 JAJ03625.1 GDIP01228120 JAI95281.1 GDIP01083296 JAM20419.1 GL888284 EGI63349.1 GDIP01109547 JAL94167.1 GDIQ01223788 JAK27937.1 GDIQ01223789 GDIQ01223787 JAK27936.1 KQ980314 KYN16692.1 GDIQ01126533 JAL25193.1 AAAB01008844 EGK96369.1 GGFK01006251 MBW39572.1 GDIQ01251280 GDIQ01251279 GDIQ01185200 GDIQ01182102 GDIQ01180798 GDIQ01163719 GDIQ01163718 GDIQ01163715 JAK70927.1 GL434548 EFN74792.1 GDIQ01242702 JAK09023.1 GDIP01108083 JAL95631.1 GDIP01228121 GDIP01219927 GDIP01174575 GDIP01158438 JAJ03475.1 KZS07507.1 GDIQ01251278 GDIQ01188299 GDIQ01185202 GDIQ01185201 GDIQ01180799 GDIQ01163717 GDIQ01163716 GDIQ01094633 JAK88009.1 GDIQ01204286 JAK47439.1 GDIQ01162366 GDIQ01111223 JAL40503.1 GDIP01177549 JAJ45853.1 GDIQ01253882 JAJ97842.1 GDIQ01216300 GDIQ01162367 GDIQ01162365 GDIQ01157523 GDIQ01112823 GDIQ01096486 GDIQ01031667 JAL55240.1 GECU01032039 JAS75667.1 GGFK01006898 MBW40219.1 GDIP01216074 JAJ07328.1 GBXI01017045 GBXI01016813 GBXI01008932 JAC97246.1 JAC97478.1 JAD05360.1 GDIP01174531 JAJ48871.1 GDIQ01248185 GDIQ01241251 GDIQ01220935 GDIQ01196405 GDIQ01177948 GDIQ01174041 GDIQ01142131 GDIQ01116533 GDIQ01115331 GDIQ01109424 GDIQ01073988 GDIQ01069503 GDIQ01069502 GDIQ01069501 GDIQ01067900 GDIQ01059899 GDIQ01057405 JAN25236.1 LBMM01005695 KMQ91287.1 GDIQ01082738 JAN11999.1 GDIQ01212735 JAK38990.1 CH916370 EDW00247.1 GDIP01028599 JAM75116.1 GDIQ01222435 GDIQ01222434 GDIQ01220936 GDIQ01220934 GDIQ01220933 GDIQ01212734 GDIQ01204243 GDIQ01186800 GDIQ01186799 GDIQ01183405 GDIQ01183404 GDIQ01183403 GDIQ01155173 GDIQ01152368 GDIQ01123933 GDIQ01067899 GDIQ01066404 GDIQ01063605 JAK68321.1 GDIQ01110931 JAL40795.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000037510
+ More
UP000092461 UP000235965 UP000078492 UP000036403 UP000245037 UP000078540 UP000008237 UP000027135 UP000005205 UP000008820 UP000078541 UP000075809 UP000092462 UP000053097 UP000009046 UP000076858 UP000000311 UP000279307 UP000069272 UP000005203 UP000007755 UP000007062 UP000075900 UP000001070
UP000092461 UP000235965 UP000078492 UP000036403 UP000245037 UP000078540 UP000008237 UP000027135 UP000005205 UP000008820 UP000078541 UP000075809 UP000092462 UP000053097 UP000009046 UP000076858 UP000000311 UP000279307 UP000069272 UP000005203 UP000007755 UP000007062 UP000075900 UP000001070
Interpro
SUPFAM
SSF51905
SSF51905
Gene 3D
ProteinModelPortal
H9JTY7
H9JTY6
H9JTY8
H9JTY5
I6YR69
A0A2A4K4A7
+ More
A0A2H1VTB0 A0A2A4K4K5 A0A3S2P1X7 A0A194PSS7 A0A194RDP5 A0A194PRT5 A0A0L7KNL7 I4DNK7 A0A194R0E8 A0A1B0GIG3 A0A1B6EWM9 A0A1B6EYD4 A0A0A9YGE8 A0A1B6HJ69 A0A1B6K5Y4 A0A2J7PXM4 A0A2J7PXQ0 A0A1B6KJY4 A0A1B6LHN9 A0A151IVV0 A0A1B6G282 A0A1B6JVY6 A0A2J7RBI7 A0A1L8E0M0 A0A1L8E037 A0A0J7K353 A0A2P8ZHS7 A0A195BBJ1 A0A1B6DKE4 E2BJJ0 A0A1L8E0E7 A0A067R8P7 A0A158NUQ9 A0A1L8DZX6 Q17DW2 A0A0P5PUH8 A0A151K093 A0A2M4ALF8 A0A0P5TWX8 A0A0P5UXE2 A0A0P5C9U0 A0A151K0E9 A0A336MWJ3 A0A151WNR9 A0A1B0D7C5 A0A026WCZ8 A0A0P5UXN7 A0A2M4AF63 E0VUB2 A0A164QA14 E2ASE9 A0A023EVK5 A0A3L8D7W0 A0A182FZT6 A0A151WMQ2 A0A0N8CY00 A0A2J7PXM0 A0A088AJU7 A0A0N7ZRH0 A0A0P4YIW2 A0A0P5WPU7 F4WR74 A0A0P5UVG6 A0A0P5JJB6 A0A0P5HR00 A0A151J323 A0A0P5Q3U3 F5HJA8 A0A2M4AFT0 A0A0P5MEE8 E1ZVM3 A0A0P5GLG5 A0A0P5UVJ4 A0A0P4ZXT5 A0A0P5LXN9 A0A0P5KVV5 A0A0P5QPX0 A0A0P5C9R7 A0A0P5FKY8 A0A0P5RT58 A0A1B6HLV3 A0A2M4AHF2 A0A0P4ZIS6 A0A0A1WEA8 A0A0P5C863 A0A0P6E0K9 A0A0J7KLX4 A0A0P6D8A7 A0A453YNJ2 A0A0P5IKE5 B4JKZ4 A0A0N8DJ56 A0A0P5M7L3 A0A0N8C666
A0A2H1VTB0 A0A2A4K4K5 A0A3S2P1X7 A0A194PSS7 A0A194RDP5 A0A194PRT5 A0A0L7KNL7 I4DNK7 A0A194R0E8 A0A1B0GIG3 A0A1B6EWM9 A0A1B6EYD4 A0A0A9YGE8 A0A1B6HJ69 A0A1B6K5Y4 A0A2J7PXM4 A0A2J7PXQ0 A0A1B6KJY4 A0A1B6LHN9 A0A151IVV0 A0A1B6G282 A0A1B6JVY6 A0A2J7RBI7 A0A1L8E0M0 A0A1L8E037 A0A0J7K353 A0A2P8ZHS7 A0A195BBJ1 A0A1B6DKE4 E2BJJ0 A0A1L8E0E7 A0A067R8P7 A0A158NUQ9 A0A1L8DZX6 Q17DW2 A0A0P5PUH8 A0A151K093 A0A2M4ALF8 A0A0P5TWX8 A0A0P5UXE2 A0A0P5C9U0 A0A151K0E9 A0A336MWJ3 A0A151WNR9 A0A1B0D7C5 A0A026WCZ8 A0A0P5UXN7 A0A2M4AF63 E0VUB2 A0A164QA14 E2ASE9 A0A023EVK5 A0A3L8D7W0 A0A182FZT6 A0A151WMQ2 A0A0N8CY00 A0A2J7PXM0 A0A088AJU7 A0A0N7ZRH0 A0A0P4YIW2 A0A0P5WPU7 F4WR74 A0A0P5UVG6 A0A0P5JJB6 A0A0P5HR00 A0A151J323 A0A0P5Q3U3 F5HJA8 A0A2M4AFT0 A0A0P5MEE8 E1ZVM3 A0A0P5GLG5 A0A0P5UVJ4 A0A0P4ZXT5 A0A0P5LXN9 A0A0P5KVV5 A0A0P5QPX0 A0A0P5C9R7 A0A0P5FKY8 A0A0P5RT58 A0A1B6HLV3 A0A2M4AHF2 A0A0P4ZIS6 A0A0A1WEA8 A0A0P5C863 A0A0P6E0K9 A0A0J7KLX4 A0A0P6D8A7 A0A453YNJ2 A0A0P5IKE5 B4JKZ4 A0A0N8DJ56 A0A0P5M7L3 A0A0N8C666
PDB
4H7U
E-value=3.33725e-49,
Score=494
Ontologies
GO
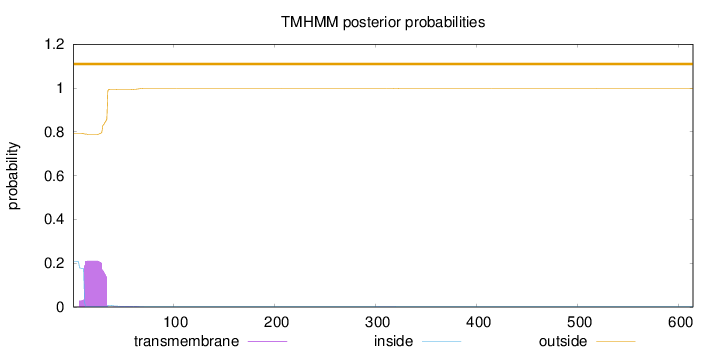
Topology
Length:
614
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.80161
Exp number, first 60 AAs:
4.7713
Total prob of N-in:
0.20716
outside
1 - 614
Population Genetic Test Statistics
Pi
265.78805
Theta
215.461299
Tajima's D
0.558225
CLR
0.241958
CSRT
0.538573071346433
Interpretation
Uncertain
