Gene
KWMTBOMO09583
Pre Gene Modal
BGIBMGA013002
Annotation
PREDICTED:_glucose_dehydrogenase_[FAD?_quinone]_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.695
Sequence
CDS
ATGGGACAAGAGCAATCTTACGTACACGCCAACGGTCCAGTGAGACGTTACAAGAACAAATACGAAGATCCTGTGCAATACAGGAACTCTATCGACTGCAGGAAAAGCGAATACGAGAGCAAGGAATACGAATACATACGACAGAAAGCGAGACGGAGCGCCGACAGTTTCAAGAGCGACCACTCCACCGAGAGCAGTGGATACCGGAGCGGATCCAGCGCCTGCGAGTGCCGATCCGAAGTCTCATCGAAAAGCGGATACTACGGCACGTCTCTGTACAAAAACGATCACTACAAAGACGCGAACAAAGACATTTACAAACCAGTGAAGCTACCAAAGGAGAAACGCTACAGGAATCAACTTATCCAACTGCAGAAGGACGAATTGAAAAGCGCCCGACTAATGAGTTATCTGGACGAAAGCGATAAAGGACAGTTTAAAACTGGGTCATCGAAAAAGTCCGGCATAAGGAATTATACTCCAAAGATTCTCTCGGAAGAGGAACAAAAGGGCAAATTGGATTGGAATATGGAGGCGGCAGGAGCTTTGGCGAGCTTGGCACCGTCGCCGATTACCGTGTTGGGTCTGATACCACTCCTGGCATTAGGGATCACGTACTTCAGATACCAACAATACGATCCGGAATCCTACGTTACGGACATCAATGCTGTGATGCCGATTTATGACTTCGTGGTAGTGGGGGGAGGTTCGGCCGGCGCTGTGGTTGCGTCGCGCCTTTCCGAGATCGGCAACTGGACCGTCCTCCTCCTCGAGGCGGGATACGACGAGAACGAGATCTCGGACATCCCCGCACTGGCGGGCTACACGCAACTCTCCGAAATGGACTGGAAGTTCCAAACGACACCTTCGAACAATCGTTCGTATTGTTTAGCCATGAACGGTGATAGATGTAATTGGCCGAGAGGTAAAGTTCTGGGTGGCAGCAGCGTACTCAATGCCATGGTATACGTTAGAGGAAACCGTAACGATTACGACTTGTGGGAGGCACTCGGTAATCCTGGATGGTCGTACGATTCCGTTCTGCCTTATTTCCTAAAATCAGAAGACAATCGGAACCCCTACTTAGTGAACACGCCGTATCACGCGGCCGGTGGATACCTGACCGTTCAAGAAGCACCATGGCGTACTCCATTATCAGTTGCGTTCTTAAAAGCCGGCATGGAACTGGGTTACGAGAACCGAGACATAAATGGAGCGAAACAGACTGGGTTCATGTTGACGCAGGCGACTATGAGGCGCGGGAGTCGATGCAGCACAGCCAAAGCGTTTCTAAGACCCGTGCGACACAGGGACAACCTTCACGTTGCGCTCGGAGCACAAGTCACCAGAATACTTATTAATCCAGTCAAAAAACAAGCATATGGGGTGGAGTTCATACGCAACGGCGTCAAACAGAAAGTCAGGATTAAGAGAGAAGTGGTAATGTCGGCCGGAGCATTGGCAACACCTCACTTGATGATGCTTAGCGGGGTTGGACCAGCGGAACACCTCAGGGAACACGGCATTCCTTTGATAGCGAATTTGAAAGTCGGCCACAACTTACAAGACCATGTCGGTTTGGGTGGATTGACCTTCGTAGTCAACAAGCCCGTCACGTTCAAGAAGGACCGATTCCAGACATTTGCTGTGGCCATGAACTACATTCTATCTGAAAAGGGACCAATGACGACCCAAGGCGTTGAAGGATTGGCTTTCGTCAATACAAAATACGCTCCGCCATCAGGGGCTTGGCCCGATATTCAGTTCCATTTCGCTCCTAGTTCAGTAAACTCTGATGGCGGAGAACAGATCCGAAAAATCTTGAATTTGCGAGATCGAGTTTACAATACCGTGTACAAACCCATTGAAAATACCGAAACCTGGACCATTCTTCCGTTACTTCTGCGCCCGAAAAGCTCCGGTTGGGTTAAATTGAGGAGTCGCAACCCTTTCCAGCCCCCCTCGATAGAGCCTAACTATTTCGCGTACAAGGAGGATATCAAAGTACTGACGGAAGGCATAAAGATCGCTATGGCGATTTCGAATACCACTTCTTTCCAAAGATACGGATCAAGGCCCCACAACATTCCCCTGCCCGGTTGCCAACACCACGTGCTCTTTAGTGACGAATACTGGGAGTGCAGTTTGAAGCACTTCTCTTTTACGATTTACCACCCCACCGGGACCTGCAAGATGGGGCCAAAACATGATCAAGAAGCCGTCGTAGACCCAAGGCTGCGAGTGCACGGTGTTGCCAACCTGCGGGTCGTGGACGCGAGTGTCATGCCGACCATTATAAGTGGCAACCCCAACGCTCCTGTCATCATGATAGCTGAGAAAGCATCGGACATGATCAAGGAGGACTGGCTTGTATTATGA
Protein
MGQEQSYVHANGPVRRYKNKYEDPVQYRNSIDCRKSEYESKEYEYIRQKARRSADSFKSDHSTESSGYRSGSSACECRSEVSSKSGYYGTSLYKNDHYKDANKDIYKPVKLPKEKRYRNQLIQLQKDELKSARLMSYLDESDKGQFKTGSSKKSGIRNYTPKILSEEEQKGKLDWNMEAAGALASLAPSPITVLGLIPLLALGITYFRYQQYDPESYVTDINAVMPIYDFVVVGGGSAGAVVASRLSEIGNWTVLLLEAGYDENEISDIPALAGYTQLSEMDWKFQTTPSNNRSYCLAMNGDRCNWPRGKVLGGSSVLNAMVYVRGNRNDYDLWEALGNPGWSYDSVLPYFLKSEDNRNPYLVNTPYHAAGGYLTVQEAPWRTPLSVAFLKAGMELGYENRDINGAKQTGFMLTQATMRRGSRCSTAKAFLRPVRHRDNLHVALGAQVTRILINPVKKQAYGVEFIRNGVKQKVRIKREVVMSAGALATPHLMMLSGVGPAEHLREHGIPLIANLKVGHNLQDHVGLGGLTFVVNKPVTFKKDRFQTFAVAMNYILSEKGPMTTQGVEGLAFVNTKYAPPSGAWPDIQFHFAPSSVNSDGGEQIRKILNLRDRVYNTVYKPIENTETWTILPLLLRPKSSGWVKLRSRNPFQPPSIEPNYFAYKEDIKVLTEGIKIAMAISNTTSFQRYGSRPHNIPLPGCQHHVLFSDEYWECSLKHFSFTIYHPTGTCKMGPKHDQEAVVDPRLRVHGVANLRVVDASVMPTIISGNPNAPVIMIAEKASDMIKEDWLVL
Summary
Cofactor
FAD
Similarity
Belongs to the GMC oxidoreductase family.
Uniprot
H9JTZ0
A0A0L7KRB9
A0A2A4JBU3
A0A2H1VYR2
A0A194QIN9
A0A194REF3
+ More
A0A212FMP7 A0A437BIL1 K7J280 A0A0M8ZQ23 A0A232EPA2 A0A310SSQ6 A0A182ITU8 Q17DW0 A0A1I8MD89 A0A182QQY2 A0A182VXW7 A0A182H3F3 A0A084VW82 A0A182MQU1 A0A1Y9HDN3 A0A453YK80 Q7PFS5 E0VUA8 A0A3B0J900 A0A182SP41 W5J3L7 A0A182G085 A0A1W4X2S9 B4L267 Q8MQY9 A0A0L0C3P5 B4JKZ5 D2A3N0 A0A154PBS2 A0A1I8PYN0 A0A034VBN3 A0A067R0L4 B4MTA0 A0A0K8UL62 A0A0C9RDB5 W8BZL2 E2ASE8 N6TYT9 A0A0A1WXX2 B3MXK4 A0A1J1IYP0 A0A0M5J614 A0A2P8ZKT8 A0A1W4WAI2 B0WJG4 A0A2J7PRB1 B3NUX6 B4M8F5 E9IKG4 A0A1A9Y1T4 A0A158NUR1 A0A1A9Z4T7 A0A1A9WC56 A0A3L8D7B4 B4R4W6 B4PW81 A0A182N4F6 A0A182JQU9 A0A0L7RJZ7 F5HJA4 A0A182VFC3 A0A182U1W7 A0A088AJV0 A0A0J7KAS9 A0A2S2QPB6 A0A1A9VSF9 A0A195BBF9 A0A2H8TYU9 F4WR73 A0A0T6BDD9 J9KB49 A0A1B0CHA7 A0A2S2N9N1 A0A1B0FCC3 U4UF26 A0A1S3D290 A0A151J2Z7 A0A151K0G8 A0A151I6J0 A0A3Q0IUJ8 E9JDH0 A0A232FGG5 K7J281 A0A151WNN3 A0A1B6DKE4 A0A0M9AA24 A0A224XLN0 A0A151J323 A0A2R7W272 A0A0L7RJJ1 F4WR74
A0A212FMP7 A0A437BIL1 K7J280 A0A0M8ZQ23 A0A232EPA2 A0A310SSQ6 A0A182ITU8 Q17DW0 A0A1I8MD89 A0A182QQY2 A0A182VXW7 A0A182H3F3 A0A084VW82 A0A182MQU1 A0A1Y9HDN3 A0A453YK80 Q7PFS5 E0VUA8 A0A3B0J900 A0A182SP41 W5J3L7 A0A182G085 A0A1W4X2S9 B4L267 Q8MQY9 A0A0L0C3P5 B4JKZ5 D2A3N0 A0A154PBS2 A0A1I8PYN0 A0A034VBN3 A0A067R0L4 B4MTA0 A0A0K8UL62 A0A0C9RDB5 W8BZL2 E2ASE8 N6TYT9 A0A0A1WXX2 B3MXK4 A0A1J1IYP0 A0A0M5J614 A0A2P8ZKT8 A0A1W4WAI2 B0WJG4 A0A2J7PRB1 B3NUX6 B4M8F5 E9IKG4 A0A1A9Y1T4 A0A158NUR1 A0A1A9Z4T7 A0A1A9WC56 A0A3L8D7B4 B4R4W6 B4PW81 A0A182N4F6 A0A182JQU9 A0A0L7RJZ7 F5HJA4 A0A182VFC3 A0A182U1W7 A0A088AJV0 A0A0J7KAS9 A0A2S2QPB6 A0A1A9VSF9 A0A195BBF9 A0A2H8TYU9 F4WR73 A0A0T6BDD9 J9KB49 A0A1B0CHA7 A0A2S2N9N1 A0A1B0FCC3 U4UF26 A0A1S3D290 A0A151J2Z7 A0A151K0G8 A0A151I6J0 A0A3Q0IUJ8 E9JDH0 A0A232FGG5 K7J281 A0A151WNN3 A0A1B6DKE4 A0A0M9AA24 A0A224XLN0 A0A151J323 A0A2R7W272 A0A0L7RJJ1 F4WR74
Pubmed
19121390
26227816
26354079
22118469
20075255
28648823
+ More
17510324 25315136 26483478 24438588 12364791 14747013 17210077 20566863 20920257 23761445 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 18362917 19820115 25348373 24845553 24495485 20798317 23537049 25830018 29403074 21282665 21347285 30249741 17550304 21719571
17510324 25315136 26483478 24438588 12364791 14747013 17210077 20566863 20920257 23761445 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 18362917 19820115 25348373 24845553 24495485 20798317 23537049 25830018 29403074 21282665 21347285 30249741 17550304 21719571
EMBL
BABH01003807
JTDY01006959
KOB65564.1
NWSH01002211
PCG68902.1
ODYU01005276
+ More
SOQ45971.1 KQ458756 KPJ05264.1 KQ460323 KPJ15967.1 AGBW02007651 OWR55008.1 RSAL01000051 RVE50267.1 AAZX01001588 KQ435998 KOX67656.1 NNAY01002979 OXU20190.1 KQ759784 OAD62857.1 CH477290 EAT44641.1 AXCN02000040 JXUM01024549 KQ560694 KXJ81237.1 ATLV01017501 KE525172 KFB42226.1 AXCM01006544 APCN01003123 AAAB01008844 EAA45201.5 DS235784 EEB16964.1 OUUW01000003 SPP78677.1 ADMH02002107 ETN58952.1 CH933810 EDW06807.2 AY122229 AE014298 AAM52741.1 AHN59721.1 JRES01000943 KNC26963.1 CH916370 EDW00248.1 KQ971348 EFA05530.2 KQ434869 KZC09325.1 GAKP01019984 GAKP01019981 GAKP01019978 GAKP01019975 JAC38968.1 KK852802 KDR16285.1 CH963851 EDW75339.2 GDHF01024877 GDHF01011981 JAI27437.1 JAI40333.1 GBYB01011122 JAG80889.1 GAMC01007801 GAMC01007799 JAB98756.1 GL442298 EFN63614.1 APGK01055694 APGK01055695 APGK01055696 APGK01055697 APGK01055698 APGK01055699 APGK01055700 APGK01055701 APGK01055702 APGK01055703 KB741269 ENN71447.1 GBXI01010590 GBXI01000279 JAD03702.1 JAD14013.1 CH902630 EDV38469.2 CVRI01000064 CRL05333.1 CP012528 ALC48844.1 PYGN01000027 PSN57080.1 DS231960 EDS29160.1 NEVH01022362 PNF18878.1 CH954180 EDV47074.1 CH940653 EDW62431.1 GL763984 EFZ18942.1 ADTU01026684 ADTU01026685 QOIP01000012 RLU16179.1 CM000366 EDX17923.1 CM000162 EDX01712.1 KQ414579 KOC71128.1 EGK96365.1 LBMM01010566 KMQ87384.1 GGMS01010167 MBY79370.1 KQ976532 KYM81562.1 GFXV01007265 MBW19070.1 GL888284 EGI63348.1 LJIG01001624 KRT85316.1 ABLF02037931 AJWK01012159 AJWK01012160 GGMR01001248 MBY13867.1 CCAG010013418 KB632330 ERL92579.1 KQ980314 KYN16691.1 KQ981296 KYN43079.1 KQ978473 KYM93711.1 GL771866 EFZ09157.1 NNAY01000267 OXU29610.1 AAZX01008029 KQ982905 KYQ49512.1 GEDC01024026 GEDC01011147 JAS13272.1 JAS26151.1 KQ435704 KOX80305.1 GFTR01007495 JAW08931.1 KYN16692.1 KK854155 PTY12285.1 KOC71127.1 EGI63349.1
SOQ45971.1 KQ458756 KPJ05264.1 KQ460323 KPJ15967.1 AGBW02007651 OWR55008.1 RSAL01000051 RVE50267.1 AAZX01001588 KQ435998 KOX67656.1 NNAY01002979 OXU20190.1 KQ759784 OAD62857.1 CH477290 EAT44641.1 AXCN02000040 JXUM01024549 KQ560694 KXJ81237.1 ATLV01017501 KE525172 KFB42226.1 AXCM01006544 APCN01003123 AAAB01008844 EAA45201.5 DS235784 EEB16964.1 OUUW01000003 SPP78677.1 ADMH02002107 ETN58952.1 CH933810 EDW06807.2 AY122229 AE014298 AAM52741.1 AHN59721.1 JRES01000943 KNC26963.1 CH916370 EDW00248.1 KQ971348 EFA05530.2 KQ434869 KZC09325.1 GAKP01019984 GAKP01019981 GAKP01019978 GAKP01019975 JAC38968.1 KK852802 KDR16285.1 CH963851 EDW75339.2 GDHF01024877 GDHF01011981 JAI27437.1 JAI40333.1 GBYB01011122 JAG80889.1 GAMC01007801 GAMC01007799 JAB98756.1 GL442298 EFN63614.1 APGK01055694 APGK01055695 APGK01055696 APGK01055697 APGK01055698 APGK01055699 APGK01055700 APGK01055701 APGK01055702 APGK01055703 KB741269 ENN71447.1 GBXI01010590 GBXI01000279 JAD03702.1 JAD14013.1 CH902630 EDV38469.2 CVRI01000064 CRL05333.1 CP012528 ALC48844.1 PYGN01000027 PSN57080.1 DS231960 EDS29160.1 NEVH01022362 PNF18878.1 CH954180 EDV47074.1 CH940653 EDW62431.1 GL763984 EFZ18942.1 ADTU01026684 ADTU01026685 QOIP01000012 RLU16179.1 CM000366 EDX17923.1 CM000162 EDX01712.1 KQ414579 KOC71128.1 EGK96365.1 LBMM01010566 KMQ87384.1 GGMS01010167 MBY79370.1 KQ976532 KYM81562.1 GFXV01007265 MBW19070.1 GL888284 EGI63348.1 LJIG01001624 KRT85316.1 ABLF02037931 AJWK01012159 AJWK01012160 GGMR01001248 MBY13867.1 CCAG010013418 KB632330 ERL92579.1 KQ980314 KYN16691.1 KQ981296 KYN43079.1 KQ978473 KYM93711.1 GL771866 EFZ09157.1 NNAY01000267 OXU29610.1 AAZX01008029 KQ982905 KYQ49512.1 GEDC01024026 GEDC01011147 JAS13272.1 JAS26151.1 KQ435704 KOX80305.1 GFTR01007495 JAW08931.1 KYN16692.1 KK854155 PTY12285.1 KOC71127.1 EGI63349.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000283053 UP000002358 UP000053105 UP000215335 UP000075880 UP000008820 UP000095301 UP000075886 UP000075920 UP000069940 UP000249989 UP000030765 UP000075883 UP000075900 UP000075840 UP000007062 UP000009046 UP000268350 UP000075901 UP000000673 UP000069272 UP000192223 UP000009192 UP000000803 UP000037069 UP000001070 UP000007266 UP000076502 UP000095300 UP000027135 UP000007798 UP000000311 UP000019118 UP000007801 UP000183832 UP000092553 UP000245037 UP000192221 UP000002320 UP000235965 UP000008711 UP000008792 UP000092443 UP000005205 UP000092445 UP000091820 UP000279307 UP000000304 UP000002282 UP000075884 UP000075881 UP000053825 UP000075903 UP000075902 UP000005203 UP000036403 UP000078200 UP000078540 UP000007755 UP000007819 UP000092461 UP000092444 UP000030742 UP000079169 UP000078492 UP000078541 UP000078542 UP000075809
UP000283053 UP000002358 UP000053105 UP000215335 UP000075880 UP000008820 UP000095301 UP000075886 UP000075920 UP000069940 UP000249989 UP000030765 UP000075883 UP000075900 UP000075840 UP000007062 UP000009046 UP000268350 UP000075901 UP000000673 UP000069272 UP000192223 UP000009192 UP000000803 UP000037069 UP000001070 UP000007266 UP000076502 UP000095300 UP000027135 UP000007798 UP000000311 UP000019118 UP000007801 UP000183832 UP000092553 UP000245037 UP000192221 UP000002320 UP000235965 UP000008711 UP000008792 UP000092443 UP000005205 UP000092445 UP000091820 UP000279307 UP000000304 UP000002282 UP000075884 UP000075881 UP000053825 UP000075903 UP000075902 UP000005203 UP000036403 UP000078200 UP000078540 UP000007755 UP000007819 UP000092461 UP000092444 UP000030742 UP000079169 UP000078492 UP000078541 UP000078542 UP000075809
Interpro
SUPFAM
SSF51905
SSF51905
Gene 3D
ProteinModelPortal
H9JTZ0
A0A0L7KRB9
A0A2A4JBU3
A0A2H1VYR2
A0A194QIN9
A0A194REF3
+ More
A0A212FMP7 A0A437BIL1 K7J280 A0A0M8ZQ23 A0A232EPA2 A0A310SSQ6 A0A182ITU8 Q17DW0 A0A1I8MD89 A0A182QQY2 A0A182VXW7 A0A182H3F3 A0A084VW82 A0A182MQU1 A0A1Y9HDN3 A0A453YK80 Q7PFS5 E0VUA8 A0A3B0J900 A0A182SP41 W5J3L7 A0A182G085 A0A1W4X2S9 B4L267 Q8MQY9 A0A0L0C3P5 B4JKZ5 D2A3N0 A0A154PBS2 A0A1I8PYN0 A0A034VBN3 A0A067R0L4 B4MTA0 A0A0K8UL62 A0A0C9RDB5 W8BZL2 E2ASE8 N6TYT9 A0A0A1WXX2 B3MXK4 A0A1J1IYP0 A0A0M5J614 A0A2P8ZKT8 A0A1W4WAI2 B0WJG4 A0A2J7PRB1 B3NUX6 B4M8F5 E9IKG4 A0A1A9Y1T4 A0A158NUR1 A0A1A9Z4T7 A0A1A9WC56 A0A3L8D7B4 B4R4W6 B4PW81 A0A182N4F6 A0A182JQU9 A0A0L7RJZ7 F5HJA4 A0A182VFC3 A0A182U1W7 A0A088AJV0 A0A0J7KAS9 A0A2S2QPB6 A0A1A9VSF9 A0A195BBF9 A0A2H8TYU9 F4WR73 A0A0T6BDD9 J9KB49 A0A1B0CHA7 A0A2S2N9N1 A0A1B0FCC3 U4UF26 A0A1S3D290 A0A151J2Z7 A0A151K0G8 A0A151I6J0 A0A3Q0IUJ8 E9JDH0 A0A232FGG5 K7J281 A0A151WNN3 A0A1B6DKE4 A0A0M9AA24 A0A224XLN0 A0A151J323 A0A2R7W272 A0A0L7RJJ1 F4WR74
A0A212FMP7 A0A437BIL1 K7J280 A0A0M8ZQ23 A0A232EPA2 A0A310SSQ6 A0A182ITU8 Q17DW0 A0A1I8MD89 A0A182QQY2 A0A182VXW7 A0A182H3F3 A0A084VW82 A0A182MQU1 A0A1Y9HDN3 A0A453YK80 Q7PFS5 E0VUA8 A0A3B0J900 A0A182SP41 W5J3L7 A0A182G085 A0A1W4X2S9 B4L267 Q8MQY9 A0A0L0C3P5 B4JKZ5 D2A3N0 A0A154PBS2 A0A1I8PYN0 A0A034VBN3 A0A067R0L4 B4MTA0 A0A0K8UL62 A0A0C9RDB5 W8BZL2 E2ASE8 N6TYT9 A0A0A1WXX2 B3MXK4 A0A1J1IYP0 A0A0M5J614 A0A2P8ZKT8 A0A1W4WAI2 B0WJG4 A0A2J7PRB1 B3NUX6 B4M8F5 E9IKG4 A0A1A9Y1T4 A0A158NUR1 A0A1A9Z4T7 A0A1A9WC56 A0A3L8D7B4 B4R4W6 B4PW81 A0A182N4F6 A0A182JQU9 A0A0L7RJZ7 F5HJA4 A0A182VFC3 A0A182U1W7 A0A088AJV0 A0A0J7KAS9 A0A2S2QPB6 A0A1A9VSF9 A0A195BBF9 A0A2H8TYU9 F4WR73 A0A0T6BDD9 J9KB49 A0A1B0CHA7 A0A2S2N9N1 A0A1B0FCC3 U4UF26 A0A1S3D290 A0A151J2Z7 A0A151K0G8 A0A151I6J0 A0A3Q0IUJ8 E9JDH0 A0A232FGG5 K7J281 A0A151WNN3 A0A1B6DKE4 A0A0M9AA24 A0A224XLN0 A0A151J323 A0A2R7W272 A0A0L7RJJ1 F4WR74
PDB
5NCC
E-value=1.19381e-66,
Score=646
Ontologies
GO
Topology
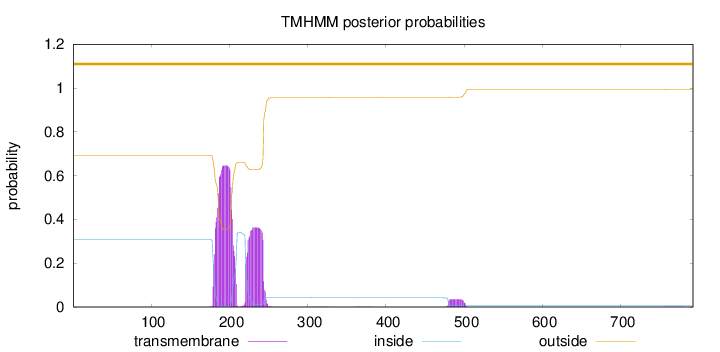
Length:
792
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
23.39486
Exp number, first 60 AAs:
0
Total prob of N-in:
0.30847
outside
1 - 792
Population Genetic Test Statistics
Pi
262.856067
Theta
176.961179
Tajima's D
0.763412
CLR
0.492819
CSRT
0.586570671466427
Interpretation
Uncertain
