Gene
KWMTBOMO09579
Pre Gene Modal
BGIBMGA013005
Annotation
PREDICTED:_glucose_dehydrogenase_[FAD?_quinone]-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.016
Sequence
CDS
ATGAGTCGCTACTGTGTGCCGAGGGCAGCGGCGGTGCTGTGGATGCTGTTCCTATTGCTGCCTGGAATAGGCACCCAGTTTGCTAACCCGATCAACTCATTACTGAACTTCGTCACTGAAGGGACTAAGCAACTCGATAACGAACCTCCTGATCAGAGGAATTTGCTGTCAGAATATGATTTCATCATAGTAGGAGCAGGCACCGCAGGGTGCGTACTTGCCAATCGACTCAGCGAAATCCAAGACTGGAAAATTTTACTCGTCGAAGCGGGCACAAACGAAAATTACCTAATGGATATTCCTATAGTAGCTAATTACCTACAATTTACAGCCGCTAATTGGGGCTACAAAACGAAACCATCTAACAAATATTGTGCTGGTTTTGAAAACAAACAATGTAACTGGCCCAGAGGAAAAGTAGTTGGAGGTTCAAGTGTACTCAATTACATGATATACACCAGGGGTTCATCAGTAGACTATAACAATTGGAAGGCGATGGGAAACGATGGCTGGGGCTGGGACGACGTACTCCCGTATTTTAAGAAAATAGAGAATTACAACATACCGTCCTTTGATGACCCAGAGTACCACGGGCACGACGGGTATTTGAATGTAGAGCATGCGCCTTTCCGCACAAAGAAAGGAAAGGCATGGGTCAAAGGAGCACAGCAATTGGGTTTTAAGTATAACGACCATAATGGAAAAAGCCCAGCAGGAGTTTCTTTCCTGCAACTGTCAATGAAAAATGGTACCCGTCACAGTTCTAGCCGAGCTTACCTGCATCCAATCAATAAAAGAAATAATCTACATTTGTCAAAGGGCAGCCTGGTGACCCGTATTATTTACGACGATGACACCAAAACTAAAGCCATAGGAATCGAATTGGAGAAATTAGGAAGAAAATATAAAATATTGGCTTCGCGTGAAGTGATTTTATCAGCGGGGGCAATAAATTCTCCACAATTGTTAATGTTATCAGGAATCGGGCCCAAGAGACATTTAGAATCAATGAACATAGAAGTCATTAAGGATTTACCTGTTGGGTATAACTTAATGGACCACATCGCTGCTGGCGGCGTGCAATTTATGGTAAACGATCAGAGTTTCTCACTGTCAACGGAGTACATATTGAGGCATTTGGAGCTGGTTTTTAAGTGGATGGCAAATCATAATGGCCCACTCTCTATTCCTGGTGGATGTGAAGCGTTGGTATTCTTGGATTTAAAGGACAAATTTAACGCAACCGGTTGGCCCGATTTGGAGTTATTGTTTATCAGTGGCGGTCTAAATTCAGACCCATTACTTCCAAGAAATTTCGGTTTCGACGAACAAATATACGCCGACACATATGGACCTTTAGGTAAAAATGATTTATTCATGGTATTTCCCATGCTGATGCGTCCCAAATCGAGAGGACGAGTGATGCTGCAAAGCAGAAATCCAAAAGCGTATCCCACTTTGATTCCTAACTATTTTGAGCACTCTGAGGATTTACAGAAGATTGTCGAAGGTATAAAAATTGCTATTGAAATTTCGAGACAACCGGCCATGAAGAAAATCGGAACAAAATTGTACGATGTACCCATAGAGGAGTGTTTGCAATACGGGCCTTTCGGAAGCGACGCATATTTTGCTTGTCAAGCGCAAATGTTCACGTTTACAATCTACCATCAAAGCGGAACTTGTAAGATGGGGCTGAAGACTGATCCGACTAGTGTCGTGGATTCAAGATTGCGTGTACACGGTATCTCTAATTTACGCGTTATAGATGCAAGCGTAATGCCAGATATAACATCGGGACATACAAACGCTCCAGTATACATGATCGCAGAGAAAGGTTCTGATATGATCAAACAAGACTGGGGTAAAATATGA
Protein
MSRYCVPRAAAVLWMLFLLLPGIGTQFANPINSLLNFVTEGTKQLDNEPPDQRNLLSEYDFIIVGAGTAGCVLANRLSEIQDWKILLVEAGTNENYLMDIPIVANYLQFTAANWGYKTKPSNKYCAGFENKQCNWPRGKVVGGSSVLNYMIYTRGSSVDYNNWKAMGNDGWGWDDVLPYFKKIENYNIPSFDDPEYHGHDGYLNVEHAPFRTKKGKAWVKGAQQLGFKYNDHNGKSPAGVSFLQLSMKNGTRHSSSRAYLHPINKRNNLHLSKGSLVTRIIYDDDTKTKAIGIELEKLGRKYKILASREVILSAGAINSPQLLMLSGIGPKRHLESMNIEVIKDLPVGYNLMDHIAAGGVQFMVNDQSFSLSTEYILRHLELVFKWMANHNGPLSIPGGCEALVFLDLKDKFNATGWPDLELLFISGGLNSDPLLPRNFGFDEQIYADTYGPLGKNDLFMVFPMLMRPKSRGRVMLQSRNPKAYPTLIPNYFEHSEDLQKIVEGIKIAIEISRQPAMKKIGTKLYDVPIEECLQYGPFGSDAYFACQAQMFTFTIYHQSGTCKMGLKTDPTSVVDSRLRVHGISNLRVIDASVMPDITSGHTNAPVYMIAEKGSDMIKQDWGKI
Summary
Cofactor
FAD
Similarity
Belongs to the GMC oxidoreductase family.
Uniprot
H9JTZ3
A0A194RET1
A0A194QP52
A0A0L7LG80
A0A291P0N5
A0A2A4JIB7
+ More
A0A291P0Y5 A0A2H1VQX3 A0A212FMU2 A0A2A4JID4 A0A1W4XEG1 D2A3N2 A0A1Y1LJW1 D2A6H9 J3JYY8 A0A2P8ZKU9 A0A067RC53 A0A1B6D9F6 A0A2J7PRD5 A0A1B6G5I8 J9JPR9 E0VUA5 A0A1B6IAY6 A0A2S2R240 A0A1B6F402 A0A1B6KBZ3 A0A2J7Q2J7 A0A2J7Q2I0 A0A2H8TE31 A0A182MA88 A0A182VXW8 A0A182H3F5 A0A453YNP4 A0A2P8YRH6 A0A182H5D0 J9K281 A0A2H8TF02 W5J5Z4 A0A182URX9 A0A2S2QDH4 A0A240PK67 A0A182U3H6 A0A182WV75 F5HJA0 A0A182I8V0 A0A182J323 B4L271 B4PW78 Q17DV8 A0A2J7PTN1 J9JJ26 A0A1B6HJF4 A0A084VGK4 B4M8G0 A0A0M4EVS3 A0A182K6F2 A0A1I8N355 B4MT97 A0A182G0C8 B4M8F9 A0A0M4EMC2 A0A084VGK5 A0A182MFV9 Q29IT0 A0A1Y9H2C8 A0A182QXU5 A0A2P8ZD27 A0A3B0K0Y7 A0A146LI23 B3MXK1 A0A1A9WC62 A0A0A9Y3M1 A0A067RKW0 B3NUX9 B4IJ69 A0A453YNL6 Q9VY10 A0A336LUV5 A0A0L0C407 A0A182QHR7 A0A336M5H8 B4JKZ2 A0A182G0C9 A0A1I8P2Q1 B4R3D2 Q29IS9 A0A182JBX2 A0A182LAT5 A0A2J7Q2K0 A0A034V5A2 A0A1A9Z4T0 A0A034V5Y1 A0A212FMT3 F5HJA1 A0A1A9VSG3 W5J8Z9 A0A1Y9H2C3 J9K6C8 A0A1A9Z4T1 A0A1W4VZR3
A0A291P0Y5 A0A2H1VQX3 A0A212FMU2 A0A2A4JID4 A0A1W4XEG1 D2A3N2 A0A1Y1LJW1 D2A6H9 J3JYY8 A0A2P8ZKU9 A0A067RC53 A0A1B6D9F6 A0A2J7PRD5 A0A1B6G5I8 J9JPR9 E0VUA5 A0A1B6IAY6 A0A2S2R240 A0A1B6F402 A0A1B6KBZ3 A0A2J7Q2J7 A0A2J7Q2I0 A0A2H8TE31 A0A182MA88 A0A182VXW8 A0A182H3F5 A0A453YNP4 A0A2P8YRH6 A0A182H5D0 J9K281 A0A2H8TF02 W5J5Z4 A0A182URX9 A0A2S2QDH4 A0A240PK67 A0A182U3H6 A0A182WV75 F5HJA0 A0A182I8V0 A0A182J323 B4L271 B4PW78 Q17DV8 A0A2J7PTN1 J9JJ26 A0A1B6HJF4 A0A084VGK4 B4M8G0 A0A0M4EVS3 A0A182K6F2 A0A1I8N355 B4MT97 A0A182G0C8 B4M8F9 A0A0M4EMC2 A0A084VGK5 A0A182MFV9 Q29IT0 A0A1Y9H2C8 A0A182QXU5 A0A2P8ZD27 A0A3B0K0Y7 A0A146LI23 B3MXK1 A0A1A9WC62 A0A0A9Y3M1 A0A067RKW0 B3NUX9 B4IJ69 A0A453YNL6 Q9VY10 A0A336LUV5 A0A0L0C407 A0A182QHR7 A0A336M5H8 B4JKZ2 A0A182G0C9 A0A1I8P2Q1 B4R3D2 Q29IS9 A0A182JBX2 A0A182LAT5 A0A2J7Q2K0 A0A034V5A2 A0A1A9Z4T0 A0A034V5Y1 A0A212FMT3 F5HJA1 A0A1A9VSG3 W5J8Z9 A0A1Y9H2C3 J9K6C8 A0A1A9Z4T1 A0A1W4VZR3
Pubmed
19121390
26354079
26227816
28986331
22118469
18362917
+ More
19820115 28004739 22516182 29403074 24845553 20566863 26483478 20920257 23761445 12364791 14747013 17210077 17994087 17550304 17510324 24438588 25315136 15632085 26823975 25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26108605 20966253 25348373
19820115 28004739 22516182 29403074 24845553 20566863 26483478 20920257 23761445 12364791 14747013 17210077 17994087 17550304 17510324 24438588 25315136 15632085 26823975 25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26108605 20966253 25348373
EMBL
BABH01003824
BABH01003825
BABH01003826
KQ460323
KPJ15969.1
KQ458756
+ More
KPJ05261.1 JTDY01001201 KOB74558.1 MF687547 ATJ44473.1 NWSH01001458 PCG71173.1 MF687602 ATJ44528.1 ODYU01003877 SOQ43186.1 AGBW02007651 OWR55010.1 PCG71172.1 KQ971348 EFA05532.1 GEZM01057608 GEZM01057607 GEZM01057606 GEZM01057605 JAV72225.1 EFA05513.1 BT128469 AEE63426.1 PYGN01000027 PSN57124.1 KK852802 KDR16287.1 GEDC01015002 GEDC01002775 JAS22296.1 JAS34523.1 NEVH01022362 PNF18879.1 GECZ01024966 GECZ01012070 JAS44803.1 JAS57699.1 ABLF02038150 ABLF02038159 ABLF02042993 DS235784 EEB16961.1 GECU01023601 JAS84105.1 GGMS01014842 MBY84045.1 GECZ01024829 JAS44940.1 GEBQ01031263 GEBQ01012250 JAT08714.1 JAT27727.1 NEVH01019370 PNF22797.1 PNF22796.1 GFXV01000539 MBW12344.1 AXCM01012634 JXUM01024564 KQ560694 KXJ81239.1 PYGN01000411 PSN46845.1 JXUM01027341 KQ560787 KXJ80882.1 ABLF02025934 GFXV01000815 MBW12620.1 ADMH02002030 ETN59862.1 GGMS01006576 MBY75779.1 AAAB01008844 EGK96361.1 APCN01003124 APCN01003125 CH933810 EDW06811.2 CM000162 EDX01709.2 CH477290 EAT44643.1 NEVH01021235 PNF19690.1 ABLF02032707 GECU01032991 JAS74715.1 ATLV01012990 KE524833 KFB37098.1 CH940653 EDW62436.2 CP012528 ALC49422.1 CH963851 EDW75336.1 EDW62435.1 ALC48846.1 ATLV01012991 KFB37099.1 CH379063 EAL32575.3 AXCN02000040 PYGN01000096 PSN54407.1 OUUW01000003 SPP78681.1 GDHC01011470 JAQ07159.1 CH902630 EDV38466.1 GBHO01015982 GBRD01010851 JAG27622.1 JAG54973.1 KK852406 KDR24506.1 CH954180 EDV47077.1 CH480847 EDW51048.1 AE014298 AAF48396.1 UFQT01000124 SSX20469.1 JRES01000943 KNC26971.1 UFQT01000589 SSX25524.1 CH916370 EDW00245.1 CM000366 EDX18503.1 EAL32574.2 PNF22812.1 GAKP01021987 JAC36965.1 GAKP01021989 JAC36963.1 OWR55009.1 EGK96362.1 ETN59863.1
KPJ05261.1 JTDY01001201 KOB74558.1 MF687547 ATJ44473.1 NWSH01001458 PCG71173.1 MF687602 ATJ44528.1 ODYU01003877 SOQ43186.1 AGBW02007651 OWR55010.1 PCG71172.1 KQ971348 EFA05532.1 GEZM01057608 GEZM01057607 GEZM01057606 GEZM01057605 JAV72225.1 EFA05513.1 BT128469 AEE63426.1 PYGN01000027 PSN57124.1 KK852802 KDR16287.1 GEDC01015002 GEDC01002775 JAS22296.1 JAS34523.1 NEVH01022362 PNF18879.1 GECZ01024966 GECZ01012070 JAS44803.1 JAS57699.1 ABLF02038150 ABLF02038159 ABLF02042993 DS235784 EEB16961.1 GECU01023601 JAS84105.1 GGMS01014842 MBY84045.1 GECZ01024829 JAS44940.1 GEBQ01031263 GEBQ01012250 JAT08714.1 JAT27727.1 NEVH01019370 PNF22797.1 PNF22796.1 GFXV01000539 MBW12344.1 AXCM01012634 JXUM01024564 KQ560694 KXJ81239.1 PYGN01000411 PSN46845.1 JXUM01027341 KQ560787 KXJ80882.1 ABLF02025934 GFXV01000815 MBW12620.1 ADMH02002030 ETN59862.1 GGMS01006576 MBY75779.1 AAAB01008844 EGK96361.1 APCN01003124 APCN01003125 CH933810 EDW06811.2 CM000162 EDX01709.2 CH477290 EAT44643.1 NEVH01021235 PNF19690.1 ABLF02032707 GECU01032991 JAS74715.1 ATLV01012990 KE524833 KFB37098.1 CH940653 EDW62436.2 CP012528 ALC49422.1 CH963851 EDW75336.1 EDW62435.1 ALC48846.1 ATLV01012991 KFB37099.1 CH379063 EAL32575.3 AXCN02000040 PYGN01000096 PSN54407.1 OUUW01000003 SPP78681.1 GDHC01011470 JAQ07159.1 CH902630 EDV38466.1 GBHO01015982 GBRD01010851 JAG27622.1 JAG54973.1 KK852406 KDR24506.1 CH954180 EDV47077.1 CH480847 EDW51048.1 AE014298 AAF48396.1 UFQT01000124 SSX20469.1 JRES01000943 KNC26971.1 UFQT01000589 SSX25524.1 CH916370 EDW00245.1 CM000366 EDX18503.1 EAL32574.2 PNF22812.1 GAKP01021987 JAC36965.1 GAKP01021989 JAC36963.1 OWR55009.1 EGK96362.1 ETN59863.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000037510
UP000218220
UP000007151
+ More
UP000192223 UP000007266 UP000245037 UP000027135 UP000235965 UP000007819 UP000009046 UP000075883 UP000075920 UP000069940 UP000249989 UP000075900 UP000000673 UP000075903 UP000075885 UP000075902 UP000076407 UP000007062 UP000075840 UP000075880 UP000009192 UP000002282 UP000008820 UP000030765 UP000008792 UP000092553 UP000075881 UP000095301 UP000007798 UP000069272 UP000001819 UP000075884 UP000075886 UP000268350 UP000007801 UP000091820 UP000008711 UP000001292 UP000000803 UP000037069 UP000001070 UP000095300 UP000000304 UP000075882 UP000092445 UP000078200 UP000192221
UP000192223 UP000007266 UP000245037 UP000027135 UP000235965 UP000007819 UP000009046 UP000075883 UP000075920 UP000069940 UP000249989 UP000075900 UP000000673 UP000075903 UP000075885 UP000075902 UP000076407 UP000007062 UP000075840 UP000075880 UP000009192 UP000002282 UP000008820 UP000030765 UP000008792 UP000092553 UP000075881 UP000095301 UP000007798 UP000069272 UP000001819 UP000075884 UP000075886 UP000268350 UP000007801 UP000091820 UP000008711 UP000001292 UP000000803 UP000037069 UP000001070 UP000095300 UP000000304 UP000075882 UP000092445 UP000078200 UP000192221
Interpro
SUPFAM
SSF51905
SSF51905
Gene 3D
ProteinModelPortal
H9JTZ3
A0A194RET1
A0A194QP52
A0A0L7LG80
A0A291P0N5
A0A2A4JIB7
+ More
A0A291P0Y5 A0A2H1VQX3 A0A212FMU2 A0A2A4JID4 A0A1W4XEG1 D2A3N2 A0A1Y1LJW1 D2A6H9 J3JYY8 A0A2P8ZKU9 A0A067RC53 A0A1B6D9F6 A0A2J7PRD5 A0A1B6G5I8 J9JPR9 E0VUA5 A0A1B6IAY6 A0A2S2R240 A0A1B6F402 A0A1B6KBZ3 A0A2J7Q2J7 A0A2J7Q2I0 A0A2H8TE31 A0A182MA88 A0A182VXW8 A0A182H3F5 A0A453YNP4 A0A2P8YRH6 A0A182H5D0 J9K281 A0A2H8TF02 W5J5Z4 A0A182URX9 A0A2S2QDH4 A0A240PK67 A0A182U3H6 A0A182WV75 F5HJA0 A0A182I8V0 A0A182J323 B4L271 B4PW78 Q17DV8 A0A2J7PTN1 J9JJ26 A0A1B6HJF4 A0A084VGK4 B4M8G0 A0A0M4EVS3 A0A182K6F2 A0A1I8N355 B4MT97 A0A182G0C8 B4M8F9 A0A0M4EMC2 A0A084VGK5 A0A182MFV9 Q29IT0 A0A1Y9H2C8 A0A182QXU5 A0A2P8ZD27 A0A3B0K0Y7 A0A146LI23 B3MXK1 A0A1A9WC62 A0A0A9Y3M1 A0A067RKW0 B3NUX9 B4IJ69 A0A453YNL6 Q9VY10 A0A336LUV5 A0A0L0C407 A0A182QHR7 A0A336M5H8 B4JKZ2 A0A182G0C9 A0A1I8P2Q1 B4R3D2 Q29IS9 A0A182JBX2 A0A182LAT5 A0A2J7Q2K0 A0A034V5A2 A0A1A9Z4T0 A0A034V5Y1 A0A212FMT3 F5HJA1 A0A1A9VSG3 W5J8Z9 A0A1Y9H2C3 J9K6C8 A0A1A9Z4T1 A0A1W4VZR3
A0A291P0Y5 A0A2H1VQX3 A0A212FMU2 A0A2A4JID4 A0A1W4XEG1 D2A3N2 A0A1Y1LJW1 D2A6H9 J3JYY8 A0A2P8ZKU9 A0A067RC53 A0A1B6D9F6 A0A2J7PRD5 A0A1B6G5I8 J9JPR9 E0VUA5 A0A1B6IAY6 A0A2S2R240 A0A1B6F402 A0A1B6KBZ3 A0A2J7Q2J7 A0A2J7Q2I0 A0A2H8TE31 A0A182MA88 A0A182VXW8 A0A182H3F5 A0A453YNP4 A0A2P8YRH6 A0A182H5D0 J9K281 A0A2H8TF02 W5J5Z4 A0A182URX9 A0A2S2QDH4 A0A240PK67 A0A182U3H6 A0A182WV75 F5HJA0 A0A182I8V0 A0A182J323 B4L271 B4PW78 Q17DV8 A0A2J7PTN1 J9JJ26 A0A1B6HJF4 A0A084VGK4 B4M8G0 A0A0M4EVS3 A0A182K6F2 A0A1I8N355 B4MT97 A0A182G0C8 B4M8F9 A0A0M4EMC2 A0A084VGK5 A0A182MFV9 Q29IT0 A0A1Y9H2C8 A0A182QXU5 A0A2P8ZD27 A0A3B0K0Y7 A0A146LI23 B3MXK1 A0A1A9WC62 A0A0A9Y3M1 A0A067RKW0 B3NUX9 B4IJ69 A0A453YNL6 Q9VY10 A0A336LUV5 A0A0L0C407 A0A182QHR7 A0A336M5H8 B4JKZ2 A0A182G0C9 A0A1I8P2Q1 B4R3D2 Q29IS9 A0A182JBX2 A0A182LAT5 A0A2J7Q2K0 A0A034V5A2 A0A1A9Z4T0 A0A034V5Y1 A0A212FMT3 F5HJA1 A0A1A9VSG3 W5J8Z9 A0A1Y9H2C3 J9K6C8 A0A1A9Z4T1 A0A1W4VZR3
PDB
4H7U
E-value=6.13063e-63,
Score=613
Ontologies
GO
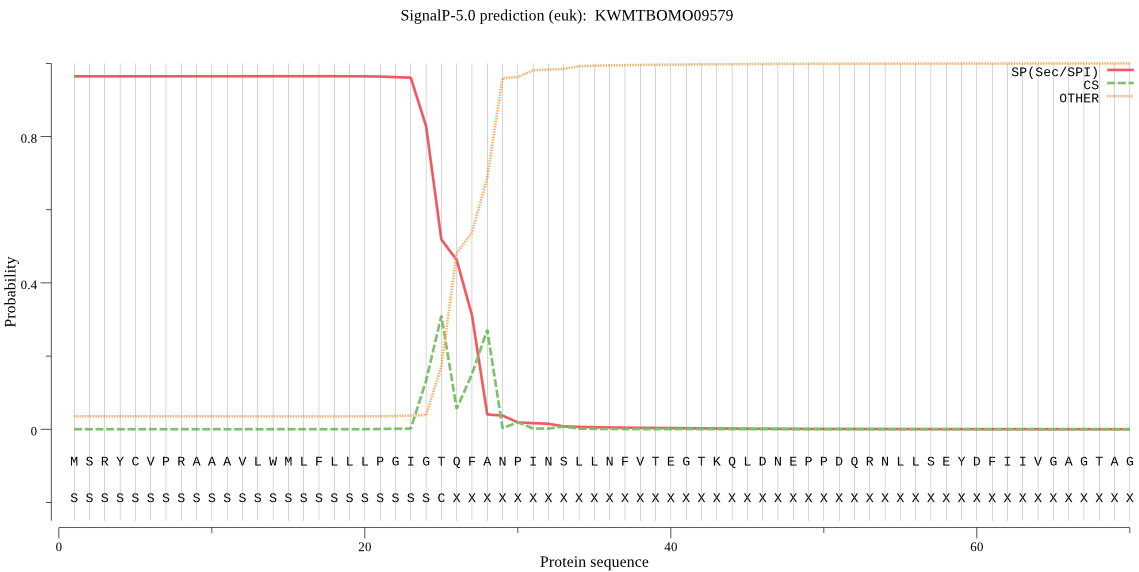
Topology
SignalP
Position: 1 - 25,
Likelihood: 0.964601
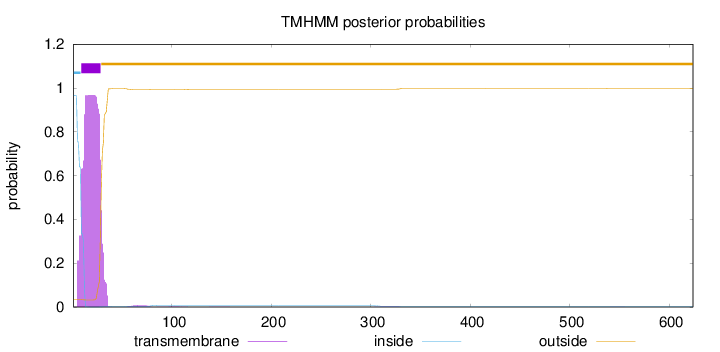
Length:
624
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.5166699999999
Exp number, first 60 AAs:
20.28461
Total prob of N-in:
0.96574
POSSIBLE N-term signal
sequence
inside
1 - 8
TMhelix
9 - 28
outside
29 - 624
Population Genetic Test Statistics
Pi
248.116036
Theta
181.143139
Tajima's D
1.234593
CLR
0.509068
CSRT
0.72386380680966
Interpretation
Uncertain
