Gene
KWMTBOMO09577 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013006
Annotation
Glucose_dehydrogenase_[acceptor]_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 1.195
Sequence
CDS
ATGTTACTGAATTATATTTCATTTTTAAGTGTTTTAATTAGCACTACAGCGTTTCATGACTACGAGTATGGCGATGATTACGAACACGATAGTGATCAATTTCGTGAACAGGGTAGATCGTATCCACAGAGTCCGATCATTGACGCAGTTTTGAAATCATACGCTCCTAATTACGTGCCGAAAAATTCTGAAGACATTTTTGATTTTTTGAGAGATTCATACCCACTGCCCGCAGGTCTTCAGAGTCCCTATGATGAATATGACTTCATTATCGTAGGGGCGGGCTCAGCAGGCTCTGTTTTAGCTTCTAGACTAACAGAAGATAAGAACACCACGGTCTTGCTTATAGAGGCCGGGAAGCCAGAAATGATATTAACTGACATTCCAATTATGGCACCATATTTTCAATCAACAGAATATGCCTGGCAGTACTATATGGAACCCCAACCTGGAGTTTGTACGGGCATGATCAACGGTCGTTGTTTTTGGCCGCGCGGGAAGGCTGTCGGGGGTAGCAGCGTCATCAACTACATGATATACACGCGTGGTAGACCACAAGAATGGGACAGAATCGCGGCTGCGGGGAATTATGGATGGTCCTATAGCGATGTTTTGGAATACTACAAAAAAGGCGAGAGAGCTAATCTAAACGGCTATGAAAAAGATCCAACGAGAAATCGCAACGGAGAAATGCCCGTTGAATTTGCACGATACAGAACCAAACTTATCGAAGCATTTTTGGAAGCGGGCAGAATCCTTGGACATCCAACAATCGATTATAATTCACCGCACCAATTAGGATTCGGTTATGTGCAAACTACAATAACCAAAGGACACCGACAAACTGCTGCGAAAGTATTTCTGCATAGACAGAAAACACGGCAAAACTTACACATCTTGCCGGACACATACGTCACTAAAATACTGATAAATCCTCAAACTAAAACCGCTTACGGAGTCGAATACGTACGCAACCGATTAAAACATAGTGTACTAGCACGTAAAGAAGTAATTCTATCTGCTGGTCCCATAGCATCCCCCCAGCTACTTATGTTATCCGGCGTGGGCCCCAAAGAACATTTAACTTCAATGGGTATACCAGTTCTCAAAGATCTGCCAGTTGGAAAAACGCTTTACGATCATATTTCTTTCCCCGCTCTTATATTCGAACTCAACACAACCGACGTCAGTTTGAACGAAAACAAGATGCTAAATGCTAAATCTGCAATTGAGTGGATTAAAAATGGTGACAATGTTATGTCATCTCCAGGGGGCGTGGAAGGAATTGGTTATATCAAAACTCCGGTCTCTAACGATCTGGATCCTATACCAGACATCGAACTGCTCAGCATTGGGGGATATATACTTGTCGATGGTGGGCCTGGAGGTAGCAAAGCGGTAAGAAAAGGAATGAGAATTAGGGATGCAGTTATAGATGAGGCTTTTGGATCAATAGACAGAATCTCTAAAGATACATGGAGTGCTTTTCCCATGTTACTACACCCAGTTTCCTTTGGAAGAATCGAACTTAAGAATAAAAATCCATTTAGTCACCCACGGATGTATGGAAATTATTTAACTCACCCAAAAGACGTGGCAACCTTCATAGCTGCAATACGTCATGTACAGGCTTTAGTTGCTACACCGCCTTTTCAGAAACTTGGTGCAAAAATACATAAAGCTCCCTACCCAACTTGCCGTGACATACCGTTTGACTCAGATGAGTACTGGGAATGCGCTATTCGCACCCTCACTGCTACTTTGCACCACCAGATTGCGACATGTCGAATGGGACCAAACAATGATTCATTAGCAGTTGTAGACCCAGAACTAAGAGTATATGGGATCGAAAAACTGCGGGTCGTAGATTCAGGAGTCATACCACGAACTATATCTGCTCATACAAATGCACCCGCGATGATGATCGCCGAAAAAGCTGCTGATATGATCAAGGCTACATGGACACATCCTTGA
Protein
MLLNYISFLSVLISTTAFHDYEYGDDYEHDSDQFREQGRSYPQSPIIDAVLKSYAPNYVPKNSEDIFDFLRDSYPLPAGLQSPYDEYDFIIVGAGSAGSVLASRLTEDKNTTVLLIEAGKPEMILTDIPIMAPYFQSTEYAWQYYMEPQPGVCTGMINGRCFWPRGKAVGGSSVINYMIYTRGRPQEWDRIAAAGNYGWSYSDVLEYYKKGERANLNGYEKDPTRNRNGEMPVEFARYRTKLIEAFLEAGRILGHPTIDYNSPHQLGFGYVQTTITKGHRQTAAKVFLHRQKTRQNLHILPDTYVTKILINPQTKTAYGVEYVRNRLKHSVLARKEVILSAGPIASPQLLMLSGVGPKEHLTSMGIPVLKDLPVGKTLYDHISFPALIFELNTTDVSLNENKMLNAKSAIEWIKNGDNVMSSPGGVEGIGYIKTPVSNDLDPIPDIELLSIGGYILVDGGPGGSKAVRKGMRIRDAVIDEAFGSIDRISKDTWSAFPMLLHPVSFGRIELKNKNPFSHPRMYGNYLTHPKDVATFIAAIRHVQALVATPPFQKLGAKIHKAPYPTCRDIPFDSDEYWECAIRTLTATLHHQIATCRMGPNNDSLAVVDPELRVYGIEKLRVVDSGVIPRTISAHTNAPAMMIAEKAADMIKATWTHP
Summary
Cofactor
FAD
Similarity
Belongs to the GMC oxidoreductase family.
Uniprot
H9JTZ4
H9JTZ5
A0A2H1W5U4
A0A437BIL7
A0A194RF91
A0A194QK24
+ More
A0A437BIJ6 A0A2A4JIB2 A0A212FMP4 A0A437BXD9 A0A212EHV5 A0A2W1BSS3 A0A194R0C6 A0A194PID4 A0A194QIN4 A0A212FMN5 A0A194RJ52 A0A0L7KWS0 H9JTZ7 A0A0L7LG80 I4DN78 A0A2H1VEA7 A0A1J1IYP0 A0A2H1V5E5 A0A336M2J5 A0A336M056 A0A1B0D7W4 E2A5J5 E9IKG7 A0A1S4F6C0 A0A139WFX6 A0A0C9PS40 Q17DV5 A0A1J1HR16 A0A0J7KKD8 B4NK50 A0A2H8TP19 J9JXJ8 A0A1S4F6D1 A0A067QS81 Q9VBG8 A0A2S2PIK7 A0A088AJU4 Q17DV6 A0A1B6DYE3 A0A182H3F8 A0A2S2PXF2 G3EIW9 B4PSV9 B4QW34 B4K6E6 F4ZNI1 F4ZNI3 K7J274 A0A182H5D1 A0A1W4VJV9 A0A182H3F7 B3LYN5 A0A154PD41 D2A3N4 G3EIW7 A0A2J7PTN1 B4GEE8 A0A310SX81 F4WR76 W4VSN1 E0VUA5 B4LYT9 A0A151I6I8 A0A2J7QH94 A0A154PDJ9 Q297A4 A0A3L8D5U9 B3P694 A0A1A9XSX2 B4JGK5 A0A182H0W9 A0A0C9RGK1 A0A0A1XHV4 A0A195BBG8 E0VUA4 A0A182GDJ6 A0A158NUV0 A0A3L8D7B5 A0A3B0KLR8 A0A0L7RJU0 B4M6M0 G3EIW8 A0A0K8U5B7 A0A1B6JGN2 A0A232FF04 A0A1A9WC53 A0A1A9Z4S3 A0A182H3F9 A0A3Q0IR20 A0A1A9VSI2 Q16P03 A0A1W4WBV5 A0A1S4FU90 A0A1W4XEG1
A0A437BIJ6 A0A2A4JIB2 A0A212FMP4 A0A437BXD9 A0A212EHV5 A0A2W1BSS3 A0A194R0C6 A0A194PID4 A0A194QIN4 A0A212FMN5 A0A194RJ52 A0A0L7KWS0 H9JTZ7 A0A0L7LG80 I4DN78 A0A2H1VEA7 A0A1J1IYP0 A0A2H1V5E5 A0A336M2J5 A0A336M056 A0A1B0D7W4 E2A5J5 E9IKG7 A0A1S4F6C0 A0A139WFX6 A0A0C9PS40 Q17DV5 A0A1J1HR16 A0A0J7KKD8 B4NK50 A0A2H8TP19 J9JXJ8 A0A1S4F6D1 A0A067QS81 Q9VBG8 A0A2S2PIK7 A0A088AJU4 Q17DV6 A0A1B6DYE3 A0A182H3F8 A0A2S2PXF2 G3EIW9 B4PSV9 B4QW34 B4K6E6 F4ZNI1 F4ZNI3 K7J274 A0A182H5D1 A0A1W4VJV9 A0A182H3F7 B3LYN5 A0A154PD41 D2A3N4 G3EIW7 A0A2J7PTN1 B4GEE8 A0A310SX81 F4WR76 W4VSN1 E0VUA5 B4LYT9 A0A151I6I8 A0A2J7QH94 A0A154PDJ9 Q297A4 A0A3L8D5U9 B3P694 A0A1A9XSX2 B4JGK5 A0A182H0W9 A0A0C9RGK1 A0A0A1XHV4 A0A195BBG8 E0VUA4 A0A182GDJ6 A0A158NUV0 A0A3L8D7B5 A0A3B0KLR8 A0A0L7RJU0 B4M6M0 G3EIW8 A0A0K8U5B7 A0A1B6JGN2 A0A232FF04 A0A1A9WC53 A0A1A9Z4S3 A0A182H3F9 A0A3Q0IR20 A0A1A9VSI2 Q16P03 A0A1W4WBV5 A0A1S4FU90 A0A1W4XEG1
Pubmed
19121390
26354079
22118469
28756777
26227816
22651552
+ More
20798317 21282665 18362917 19820115 17510324 17994087 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26483478 21429930 17550304 18057021 21383196 20075255 21719571 24391959 20566863 15632085 23185243 30249741 25830018 21347285 28648823
20798317 21282665 18362917 19820115 17510324 17994087 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26483478 21429930 17550304 18057021 21383196 20075255 21719571 24391959 20566863 15632085 23185243 30249741 25830018 21347285 28648823
EMBL
BABH01003829
BABH01003830
BABH01003831
ODYU01006526
SOQ48459.1
RSAL01000051
+ More
RVE50271.1 KQ460323 KPJ15970.1 KQ458756 KPJ05260.1 RVE50270.1 NWSH01001458 PCG71170.1 AGBW02007651 OWR55011.1 RSAL01000001 RVE55122.1 AGBW02014789 OWR41062.1 KZ149911 PZC78108.1 KQ460883 KPJ11152.1 KQ459603 KPI92808.1 KPJ05259.1 OWR55012.1 KPJ15971.1 JTDY01004843 KOB67682.1 BABH01003834 JTDY01001201 KOB74558.1 AK402758 BAM19368.1 ODYU01002091 SOQ39165.1 CVRI01000064 CRL05333.1 ODYU01000745 SOQ35996.1 UFQS01000369 UFQT01000369 SSX03245.1 SSX23611.1 SSX03246.1 SSX23612.1 AJVK01032026 GL436992 EFN71295.1 GL763984 EFZ18940.1 KQ971348 KYB26873.1 GBYB01004178 JAG73945.1 CH477290 EAT44646.1 CVRI01000019 CRK90469.1 LBMM01006334 KMQ90686.1 CH964272 EDW85092.1 GFXV01004112 MBW15917.1 ABLF02037922 KK853018 KDR12457.1 AE014297 AAF56574.1 GGMR01016673 MBY29292.1 EAT44645.1 GEDC01006623 JAS30675.1 JXUM01024565 KQ560694 KXJ81242.1 GGMS01001005 MBY70208.1 HQ857158 AEM60160.1 CM000160 EDW98646.1 CM000364 EDX14533.1 CH933806 EDW15219.1 KRG01446.1 HQ245150 AEB91339.1 HQ245152 AEB91341.1 AAZX01003729 JXUM01027342 KQ560787 KXJ80883.1 JXUM01024564 KXJ81241.1 CH902617 EDV42950.1 KQ434869 KZC09314.1 EFA05534.2 HQ857156 AEM60158.1 NEVH01021235 PNF19690.1 CH479182 EDW33983.1 KQ759784 OAD62863.1 GL888284 EGI63351.1 GAPU01000071 JAB84774.1 DS235784 EEB16961.1 CH940650 EDW67016.1 KRF83055.1 KQ978473 KYM93709.1 NEVH01013984 PNF27957.1 KZC09328.1 CM000070 EAL28302.2 KRT00640.1 QOIP01000012 RLU15835.1 CH954182 EDV53564.1 CH916369 EDV93702.1 JXUM01102122 JXUM01102123 JXUM01102124 JXUM01102125 KQ564702 KXJ71872.1 GBYB01007495 JAG77262.1 GBXI01003797 JAD10495.1 KQ976532 KYM81560.1 EEB16960.1 JXUM01056073 JXUM01056074 KQ561895 KXJ77234.1 ADTU01026685 RLU16174.1 OUUW01000023 SPP89560.1 KQ414579 KOC71135.1 CH940653 EDW62437.2 HQ857157 AEM60159.1 GDHF01030739 JAI21575.1 GECU01009357 JAS98349.1 NNAY01000314 OXU29282.1 KXJ81243.1 CH477801 EAT36071.1
RVE50271.1 KQ460323 KPJ15970.1 KQ458756 KPJ05260.1 RVE50270.1 NWSH01001458 PCG71170.1 AGBW02007651 OWR55011.1 RSAL01000001 RVE55122.1 AGBW02014789 OWR41062.1 KZ149911 PZC78108.1 KQ460883 KPJ11152.1 KQ459603 KPI92808.1 KPJ05259.1 OWR55012.1 KPJ15971.1 JTDY01004843 KOB67682.1 BABH01003834 JTDY01001201 KOB74558.1 AK402758 BAM19368.1 ODYU01002091 SOQ39165.1 CVRI01000064 CRL05333.1 ODYU01000745 SOQ35996.1 UFQS01000369 UFQT01000369 SSX03245.1 SSX23611.1 SSX03246.1 SSX23612.1 AJVK01032026 GL436992 EFN71295.1 GL763984 EFZ18940.1 KQ971348 KYB26873.1 GBYB01004178 JAG73945.1 CH477290 EAT44646.1 CVRI01000019 CRK90469.1 LBMM01006334 KMQ90686.1 CH964272 EDW85092.1 GFXV01004112 MBW15917.1 ABLF02037922 KK853018 KDR12457.1 AE014297 AAF56574.1 GGMR01016673 MBY29292.1 EAT44645.1 GEDC01006623 JAS30675.1 JXUM01024565 KQ560694 KXJ81242.1 GGMS01001005 MBY70208.1 HQ857158 AEM60160.1 CM000160 EDW98646.1 CM000364 EDX14533.1 CH933806 EDW15219.1 KRG01446.1 HQ245150 AEB91339.1 HQ245152 AEB91341.1 AAZX01003729 JXUM01027342 KQ560787 KXJ80883.1 JXUM01024564 KXJ81241.1 CH902617 EDV42950.1 KQ434869 KZC09314.1 EFA05534.2 HQ857156 AEM60158.1 NEVH01021235 PNF19690.1 CH479182 EDW33983.1 KQ759784 OAD62863.1 GL888284 EGI63351.1 GAPU01000071 JAB84774.1 DS235784 EEB16961.1 CH940650 EDW67016.1 KRF83055.1 KQ978473 KYM93709.1 NEVH01013984 PNF27957.1 KZC09328.1 CM000070 EAL28302.2 KRT00640.1 QOIP01000012 RLU15835.1 CH954182 EDV53564.1 CH916369 EDV93702.1 JXUM01102122 JXUM01102123 JXUM01102124 JXUM01102125 KQ564702 KXJ71872.1 GBYB01007495 JAG77262.1 GBXI01003797 JAD10495.1 KQ976532 KYM81560.1 EEB16960.1 JXUM01056073 JXUM01056074 KQ561895 KXJ77234.1 ADTU01026685 RLU16174.1 OUUW01000023 SPP89560.1 KQ414579 KOC71135.1 CH940653 EDW62437.2 HQ857157 AEM60159.1 GDHF01030739 JAI21575.1 GECU01009357 JAS98349.1 NNAY01000314 OXU29282.1 KXJ81243.1 CH477801 EAT36071.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000053268
UP000218220
UP000007151
+ More
UP000037510 UP000183832 UP000092462 UP000000311 UP000007266 UP000008820 UP000036403 UP000007798 UP000007819 UP000027135 UP000000803 UP000005203 UP000069940 UP000249989 UP000002282 UP000000304 UP000009192 UP000002358 UP000192221 UP000007801 UP000076502 UP000235965 UP000008744 UP000007755 UP000009046 UP000008792 UP000078542 UP000001819 UP000279307 UP000008711 UP000092443 UP000001070 UP000078540 UP000005205 UP000268350 UP000053825 UP000215335 UP000091820 UP000092445 UP000079169 UP000078200 UP000192223
UP000037510 UP000183832 UP000092462 UP000000311 UP000007266 UP000008820 UP000036403 UP000007798 UP000007819 UP000027135 UP000000803 UP000005203 UP000069940 UP000249989 UP000002282 UP000000304 UP000009192 UP000002358 UP000192221 UP000007801 UP000076502 UP000235965 UP000008744 UP000007755 UP000009046 UP000008792 UP000078542 UP000001819 UP000279307 UP000008711 UP000092443 UP000001070 UP000078540 UP000005205 UP000268350 UP000053825 UP000215335 UP000091820 UP000092445 UP000079169 UP000078200 UP000192223
Interpro
SUPFAM
SSF51905
SSF51905
Gene 3D
ProteinModelPortal
H9JTZ4
H9JTZ5
A0A2H1W5U4
A0A437BIL7
A0A194RF91
A0A194QK24
+ More
A0A437BIJ6 A0A2A4JIB2 A0A212FMP4 A0A437BXD9 A0A212EHV5 A0A2W1BSS3 A0A194R0C6 A0A194PID4 A0A194QIN4 A0A212FMN5 A0A194RJ52 A0A0L7KWS0 H9JTZ7 A0A0L7LG80 I4DN78 A0A2H1VEA7 A0A1J1IYP0 A0A2H1V5E5 A0A336M2J5 A0A336M056 A0A1B0D7W4 E2A5J5 E9IKG7 A0A1S4F6C0 A0A139WFX6 A0A0C9PS40 Q17DV5 A0A1J1HR16 A0A0J7KKD8 B4NK50 A0A2H8TP19 J9JXJ8 A0A1S4F6D1 A0A067QS81 Q9VBG8 A0A2S2PIK7 A0A088AJU4 Q17DV6 A0A1B6DYE3 A0A182H3F8 A0A2S2PXF2 G3EIW9 B4PSV9 B4QW34 B4K6E6 F4ZNI1 F4ZNI3 K7J274 A0A182H5D1 A0A1W4VJV9 A0A182H3F7 B3LYN5 A0A154PD41 D2A3N4 G3EIW7 A0A2J7PTN1 B4GEE8 A0A310SX81 F4WR76 W4VSN1 E0VUA5 B4LYT9 A0A151I6I8 A0A2J7QH94 A0A154PDJ9 Q297A4 A0A3L8D5U9 B3P694 A0A1A9XSX2 B4JGK5 A0A182H0W9 A0A0C9RGK1 A0A0A1XHV4 A0A195BBG8 E0VUA4 A0A182GDJ6 A0A158NUV0 A0A3L8D7B5 A0A3B0KLR8 A0A0L7RJU0 B4M6M0 G3EIW8 A0A0K8U5B7 A0A1B6JGN2 A0A232FF04 A0A1A9WC53 A0A1A9Z4S3 A0A182H3F9 A0A3Q0IR20 A0A1A9VSI2 Q16P03 A0A1W4WBV5 A0A1S4FU90 A0A1W4XEG1
A0A437BIJ6 A0A2A4JIB2 A0A212FMP4 A0A437BXD9 A0A212EHV5 A0A2W1BSS3 A0A194R0C6 A0A194PID4 A0A194QIN4 A0A212FMN5 A0A194RJ52 A0A0L7KWS0 H9JTZ7 A0A0L7LG80 I4DN78 A0A2H1VEA7 A0A1J1IYP0 A0A2H1V5E5 A0A336M2J5 A0A336M056 A0A1B0D7W4 E2A5J5 E9IKG7 A0A1S4F6C0 A0A139WFX6 A0A0C9PS40 Q17DV5 A0A1J1HR16 A0A0J7KKD8 B4NK50 A0A2H8TP19 J9JXJ8 A0A1S4F6D1 A0A067QS81 Q9VBG8 A0A2S2PIK7 A0A088AJU4 Q17DV6 A0A1B6DYE3 A0A182H3F8 A0A2S2PXF2 G3EIW9 B4PSV9 B4QW34 B4K6E6 F4ZNI1 F4ZNI3 K7J274 A0A182H5D1 A0A1W4VJV9 A0A182H3F7 B3LYN5 A0A154PD41 D2A3N4 G3EIW7 A0A2J7PTN1 B4GEE8 A0A310SX81 F4WR76 W4VSN1 E0VUA5 B4LYT9 A0A151I6I8 A0A2J7QH94 A0A154PDJ9 Q297A4 A0A3L8D5U9 B3P694 A0A1A9XSX2 B4JGK5 A0A182H0W9 A0A0C9RGK1 A0A0A1XHV4 A0A195BBG8 E0VUA4 A0A182GDJ6 A0A158NUV0 A0A3L8D7B5 A0A3B0KLR8 A0A0L7RJU0 B4M6M0 G3EIW8 A0A0K8U5B7 A0A1B6JGN2 A0A232FF04 A0A1A9WC53 A0A1A9Z4S3 A0A182H3F9 A0A3Q0IR20 A0A1A9VSI2 Q16P03 A0A1W4WBV5 A0A1S4FU90 A0A1W4XEG1
PDB
5OC1
E-value=1.32784e-56,
Score=558
Ontologies
GO
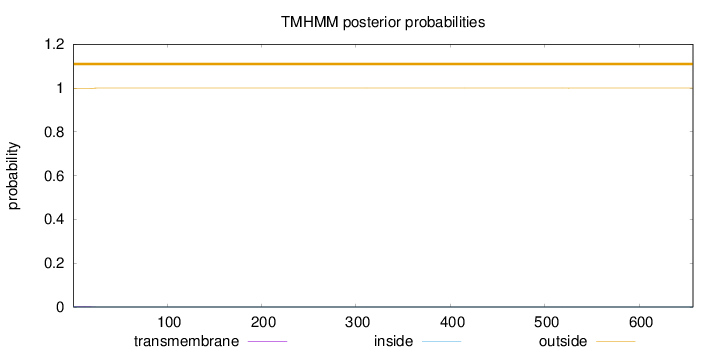
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.692933
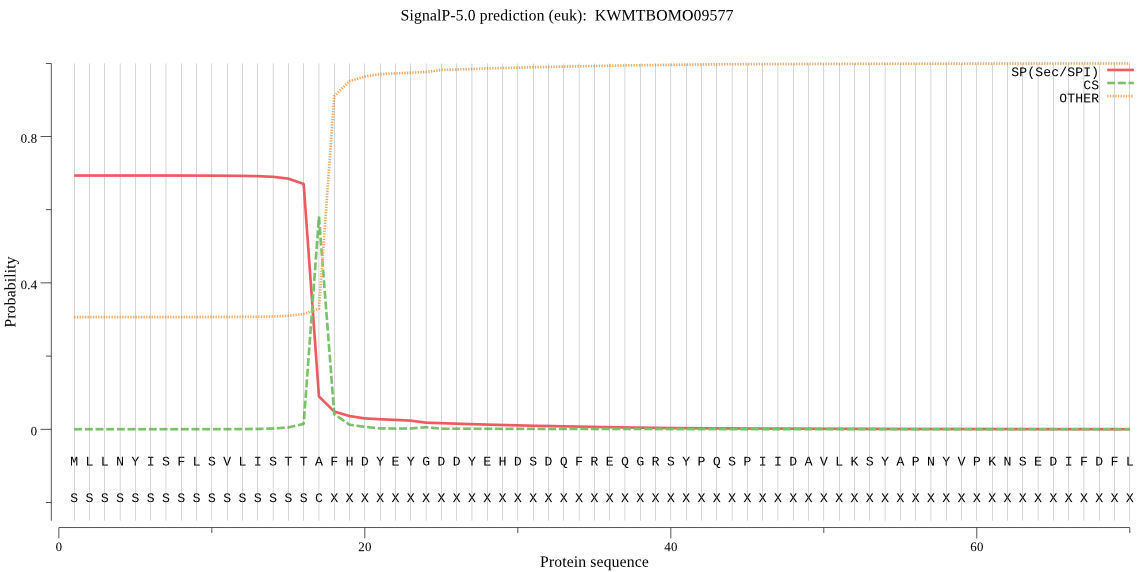
Length:
657
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0621600000000002
Exp number, first 60 AAs:
0.05799
Total prob of N-in:
0.00317
outside
1 - 657
Population Genetic Test Statistics
Pi
23.568863
Theta
203.661888
Tajima's D
0.355249
CLR
0.615526
CSRT
0.472876356182191
Interpretation
Uncertain
