Gene
KWMTBOMO09575 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013007
Annotation
PREDICTED:_glucose_dehydrogenase_[FAD?_quinone]-like_isoform_X1_[Papilio_machaon]
Location in the cell
Mitochondrial Reliability : 3.316
Sequence
CDS
ATGCAATTGACAGTAAATTTACTCAATGGTTATGAAAAAGATCCAACAAGGAATCGTAATGGGGAAAACCCTGTGGAATTTGTACCGATCAGGACTAAGCTCATCAAAGCATTTTTAGAGGCCGGTAGAATCTTTGGACATCCAACAATTGATTACAATTCACCGCACCAATTAGGATTCGGTTATGTGCAAGCTATAACAACCAAAGGACACCGACAAAGTGCTGCGAAAGTATTCCTGCATAGACAGAAGAAGCGACAAAACTTACACATCTTGCCGGACACATACGTCACTAAAATACTCATAGATGCTCGAACTAAGACAGCCTATGGAGTCGAATACGTACGAAACAGATTAAAACACACTGTACTAGCACGAAAAGAAGTAATTCTATCAGCCGGTCCCATAGCGTCTCCTCAGCTACTTATGTTATCCGGCGTGGGGCCTAAAAAACACTTAAATTCAATGGGCATACCGGTTCTCAAAGATCTGCCAGTTGGTAAAATGCTATACGATCATATTTGTTTCCCGGGACTTATATTTGAACTCAACACAACCGATGTCAGCTTGCACGAAAGCAAAATGTTAAGAGTGCGACCTATACTGCAGTGGTTAAAGAATGGTGATAGTACACTATCATCTCCGGGTGGCGTGGAAGGAATCGGTTATATCAAAACTCCGATCTCCAACGATCCAGATCCTATACCCGATATTGAACTAATCAGTATCGGTGGATCTATACTGTCAGATGGTGGGCCTGGAGGCAGTAGAGCAATTCGAAGAGGAATGAGAATTAGGGATGCAGTTATAGATGAGGCTTTTGGATCAATAGATAGAGTCGCAAGAGATACTTGGAGTGCTTTTATTTTGTTGCTACACCCAGTTTCATTTGGGGGAATAGAGCTTAAAAATAAAAACCCATTTAGTCATCCCCGGATGTATGGAAATTATTTAACTCATTCGAAGGATGTAGTAACAATTATTGCTGGGATACGATACGTACAGGCTATGGCGGCAACATTCCCTTTCCAGAAGTTTGGTGCAAAAGTACACAAAGCGTCCTATCCAACTTGCCGACACATACCGTTTGATTCAGATGAGTACTGGGAATGCGCTATTCGCACCCTCACTGCTACTTTGCACCACCAGATTGCGACGTGTCGAATGGGACCAAACAATGATCCATTAGCAGTTGTAGACCCAGAACTAAGAGTATATGGGATCGAAAAACTGCGGGTCGTAGATTCAGGAGTCATACCACGAACTATATCTGCTCATACACATGCACCCACGATGATGATCGCTGAAAAAGCTGCTGATATGATCAAAGCTACATGGGCATCAATGCTACCTAATTACATTTCCTATTCATAA
Protein
MQLTVNLLNGYEKDPTRNRNGENPVEFVPIRTKLIKAFLEAGRIFGHPTIDYNSPHQLGFGYVQAITTKGHRQSAAKVFLHRQKKRQNLHILPDTYVTKILIDARTKTAYGVEYVRNRLKHTVLARKEVILSAGPIASPQLLMLSGVGPKKHLNSMGIPVLKDLPVGKMLYDHICFPGLIFELNTTDVSLHESKMLRVRPILQWLKNGDSTLSSPGGVEGIGYIKTPISNDPDPIPDIELISIGGSILSDGGPGGSRAIRRGMRIRDAVIDEAFGSIDRVARDTWSAFILLLHPVSFGGIELKNKNPFSHPRMYGNYLTHSKDVVTIIAGIRYVQAMAATFPFQKFGAKVHKASYPTCRHIPFDSDEYWECAIRTLTATLHHQIATCRMGPNNDPLAVVDPELRVYGIEKLRVVDSGVIPRTISAHTHAPTMMIAEKAADMIKATWASMLPNYISYS
Summary
Cofactor
FAD
Similarity
Belongs to the GMC oxidoreductase family.
Uniprot
H9JTZ5
H9JTZ4
A0A2H1W5U4
A0A194RF91
A0A194QK24
A0A437BIL7
+ More
A0A2A4JIB2 A0A437BIJ6 A0A212FMP4 A0A0L7LG80 A0A437BXD9 A0A0L7KWS0 A0A2W1BSS3 A0A212EHV5 I4DN78 A0A194QIN4 A0A212FMN5 A0A194RJ52 A0A194PID4 A0A194R0C6 A0A2H1VEA7 H9JTZ7 A0A0L7LTW1 A0A1B0D7W4 A0A1J1IYP0 A0A1S4F6D1 A0A182H3F7 D2A3N4 Q17DV6 G3EIW9 A0A336M056 A0A336M2J5 A0A182VDS3 A0A182WV76 A0A182LAT9 A0A182I8V1 Q7PFS6 E2A5J5 A0A2J7QH94 A0A182SY41 A0A182TTM6 F4ZNH7 A0A067QS81 A0A182VXW9 A0A182MAY6 E0VG36 A0A0M9AA53 A0A182R5V2 A0A182K6F1 A0A139WFX6 Q17DV5 A0A154PD41 A0A084VGK6 A0A1J1HR16 A0A182IZU5 A0A0A9VXY2 A0A088AJU4 A0A3L8D5U9 G3EIW8 B4MT95 A0A182H3F8 A0A1I8NUQ1 A0A2A3EHS4 A0A1S4F6C0 A0A158NUU8 G3EIW7 A0A310SX81 B4JKZ1 Q29IT2 A0A195BAK2 K7J0B9 A0A182F7P4 A0A2S2PXF2 A0A182H5D1 A0A195E3M0 A0A195FTT2 A0A1A9XSX2 A0A0L7RJU0 A0A182LZG9 W5JAQ1 B4M6M0 E2BJJ7 B4GEE8 E1ZVM3 A0A182U7N2 A0A182JLA4 A0A3B0JDI1 A0A182HSQ7 F4ZNH5 A0A232EID2 B4QW34 A0A2H1V5E5 A0A182UYY5 Q297A4 F4WAD9 A0A1S4GF89 A0A182LES8 Q7QCC0 A0A0L7L1A4 B4HAM2
A0A2A4JIB2 A0A437BIJ6 A0A212FMP4 A0A0L7LG80 A0A437BXD9 A0A0L7KWS0 A0A2W1BSS3 A0A212EHV5 I4DN78 A0A194QIN4 A0A212FMN5 A0A194RJ52 A0A194PID4 A0A194R0C6 A0A2H1VEA7 H9JTZ7 A0A0L7LTW1 A0A1B0D7W4 A0A1J1IYP0 A0A1S4F6D1 A0A182H3F7 D2A3N4 Q17DV6 G3EIW9 A0A336M056 A0A336M2J5 A0A182VDS3 A0A182WV76 A0A182LAT9 A0A182I8V1 Q7PFS6 E2A5J5 A0A2J7QH94 A0A182SY41 A0A182TTM6 F4ZNH7 A0A067QS81 A0A182VXW9 A0A182MAY6 E0VG36 A0A0M9AA53 A0A182R5V2 A0A182K6F1 A0A139WFX6 Q17DV5 A0A154PD41 A0A084VGK6 A0A1J1HR16 A0A182IZU5 A0A0A9VXY2 A0A088AJU4 A0A3L8D5U9 G3EIW8 B4MT95 A0A182H3F8 A0A1I8NUQ1 A0A2A3EHS4 A0A1S4F6C0 A0A158NUU8 G3EIW7 A0A310SX81 B4JKZ1 Q29IT2 A0A195BAK2 K7J0B9 A0A182F7P4 A0A2S2PXF2 A0A182H5D1 A0A195E3M0 A0A195FTT2 A0A1A9XSX2 A0A0L7RJU0 A0A182LZG9 W5JAQ1 B4M6M0 E2BJJ7 B4GEE8 E1ZVM3 A0A182U7N2 A0A182JLA4 A0A3B0JDI1 A0A182HSQ7 F4ZNH5 A0A232EID2 B4QW34 A0A2H1V5E5 A0A182UYY5 Q297A4 F4WAD9 A0A1S4GF89 A0A182LES8 Q7QCC0 A0A0L7L1A4 B4HAM2
Pubmed
EMBL
BABH01003831
BABH01003829
BABH01003830
ODYU01006526
SOQ48459.1
KQ460323
+ More
KPJ15970.1 KQ458756 KPJ05260.1 RSAL01000051 RVE50271.1 NWSH01001458 PCG71170.1 RVE50270.1 AGBW02007651 OWR55011.1 JTDY01001201 KOB74558.1 RSAL01000001 RVE55122.1 JTDY01004843 KOB67682.1 KZ149911 PZC78108.1 AGBW02014789 OWR41062.1 AK402758 BAM19368.1 KPJ05259.1 OWR55012.1 KPJ15971.1 KQ459603 KPI92808.1 KQ460883 KPJ11152.1 ODYU01002091 SOQ39165.1 BABH01003834 JTDY01000138 KOB78646.1 AJVK01032026 CVRI01000064 CRL05333.1 JXUM01024564 KQ560694 KXJ81241.1 KQ971348 EFA05534.2 CH477290 EAT44645.1 HQ857158 AEM60160.1 UFQS01000369 UFQT01000369 SSX03246.1 SSX23612.1 SSX03245.1 SSX23611.1 APCN01003125 AAAB01008844 EAA45200.5 GL436992 EFN71295.1 NEVH01013984 PNF27957.1 HQ245146 AEB91347.1 KK853018 KDR12457.1 AXCM01002814 DS235131 EEB12342.1 KQ435703 KOX80330.1 KYB26873.1 EAT44646.1 KQ434869 KZC09314.1 ATLV01012991 KE524833 KFB37100.1 CVRI01000019 CRK90469.1 GBHO01043105 JAG00499.1 QOIP01000012 RLU15835.1 HQ857157 AEM60159.1 CH963851 EDW75334.1 JXUM01024565 KXJ81242.1 KZ288238 PBC31353.1 ADTU01026680 HQ857156 AEM60158.1 KQ759784 OAD62863.1 CH916370 EDW00244.1 CH379063 EAL32571.3 KQ976532 KYM81568.1 GGMS01001005 MBY70208.1 JXUM01027342 KQ560787 KXJ80883.1 KQ979701 KYN19666.1 KQ981281 KYN43299.1 KQ414579 KOC71135.1 AXCM01005241 ADMH02002030 ETN59864.1 CH940653 EDW62437.2 GL448571 EFN84154.1 CH479182 EDW33983.1 GL434548 EFN74792.1 OUUW01000003 SPP78683.1 APCN01000325 HQ245144 AEB91345.1 NNAY01004263 OXU18129.1 CM000364 EDX14533.1 ODYU01000745 SOQ35996.1 CM000070 EAL28302.2 KRT00640.1 GL888048 EGI68822.1 AAAB01008859 EAA07534.5 JTDY01003720 KOB69096.1 CH479242 EDW37633.1
KPJ15970.1 KQ458756 KPJ05260.1 RSAL01000051 RVE50271.1 NWSH01001458 PCG71170.1 RVE50270.1 AGBW02007651 OWR55011.1 JTDY01001201 KOB74558.1 RSAL01000001 RVE55122.1 JTDY01004843 KOB67682.1 KZ149911 PZC78108.1 AGBW02014789 OWR41062.1 AK402758 BAM19368.1 KPJ05259.1 OWR55012.1 KPJ15971.1 KQ459603 KPI92808.1 KQ460883 KPJ11152.1 ODYU01002091 SOQ39165.1 BABH01003834 JTDY01000138 KOB78646.1 AJVK01032026 CVRI01000064 CRL05333.1 JXUM01024564 KQ560694 KXJ81241.1 KQ971348 EFA05534.2 CH477290 EAT44645.1 HQ857158 AEM60160.1 UFQS01000369 UFQT01000369 SSX03246.1 SSX23612.1 SSX03245.1 SSX23611.1 APCN01003125 AAAB01008844 EAA45200.5 GL436992 EFN71295.1 NEVH01013984 PNF27957.1 HQ245146 AEB91347.1 KK853018 KDR12457.1 AXCM01002814 DS235131 EEB12342.1 KQ435703 KOX80330.1 KYB26873.1 EAT44646.1 KQ434869 KZC09314.1 ATLV01012991 KE524833 KFB37100.1 CVRI01000019 CRK90469.1 GBHO01043105 JAG00499.1 QOIP01000012 RLU15835.1 HQ857157 AEM60159.1 CH963851 EDW75334.1 JXUM01024565 KXJ81242.1 KZ288238 PBC31353.1 ADTU01026680 HQ857156 AEM60158.1 KQ759784 OAD62863.1 CH916370 EDW00244.1 CH379063 EAL32571.3 KQ976532 KYM81568.1 GGMS01001005 MBY70208.1 JXUM01027342 KQ560787 KXJ80883.1 KQ979701 KYN19666.1 KQ981281 KYN43299.1 KQ414579 KOC71135.1 AXCM01005241 ADMH02002030 ETN59864.1 CH940653 EDW62437.2 GL448571 EFN84154.1 CH479182 EDW33983.1 GL434548 EFN74792.1 OUUW01000003 SPP78683.1 APCN01000325 HQ245144 AEB91345.1 NNAY01004263 OXU18129.1 CM000364 EDX14533.1 ODYU01000745 SOQ35996.1 CM000070 EAL28302.2 KRT00640.1 GL888048 EGI68822.1 AAAB01008859 EAA07534.5 JTDY01003720 KOB69096.1 CH479242 EDW37633.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000218220
UP000007151
+ More
UP000037510 UP000092462 UP000183832 UP000069940 UP000249989 UP000007266 UP000008820 UP000075903 UP000076407 UP000075882 UP000075840 UP000007062 UP000000311 UP000235965 UP000075901 UP000075902 UP000027135 UP000075920 UP000075883 UP000009046 UP000053105 UP000075900 UP000075881 UP000076502 UP000030765 UP000075880 UP000005203 UP000279307 UP000007798 UP000095300 UP000242457 UP000005205 UP000001070 UP000001819 UP000078540 UP000002358 UP000069272 UP000078492 UP000078541 UP000092443 UP000053825 UP000000673 UP000008792 UP000008237 UP000008744 UP000268350 UP000215335 UP000000304 UP000007755
UP000037510 UP000092462 UP000183832 UP000069940 UP000249989 UP000007266 UP000008820 UP000075903 UP000076407 UP000075882 UP000075840 UP000007062 UP000000311 UP000235965 UP000075901 UP000075902 UP000027135 UP000075920 UP000075883 UP000009046 UP000053105 UP000075900 UP000075881 UP000076502 UP000030765 UP000075880 UP000005203 UP000279307 UP000007798 UP000095300 UP000242457 UP000005205 UP000001070 UP000001819 UP000078540 UP000002358 UP000069272 UP000078492 UP000078541 UP000092443 UP000053825 UP000000673 UP000008792 UP000008237 UP000008744 UP000268350 UP000215335 UP000000304 UP000007755
Interpro
SUPFAM
SSF51905
SSF51905
Gene 3D
ProteinModelPortal
H9JTZ5
H9JTZ4
A0A2H1W5U4
A0A194RF91
A0A194QK24
A0A437BIL7
+ More
A0A2A4JIB2 A0A437BIJ6 A0A212FMP4 A0A0L7LG80 A0A437BXD9 A0A0L7KWS0 A0A2W1BSS3 A0A212EHV5 I4DN78 A0A194QIN4 A0A212FMN5 A0A194RJ52 A0A194PID4 A0A194R0C6 A0A2H1VEA7 H9JTZ7 A0A0L7LTW1 A0A1B0D7W4 A0A1J1IYP0 A0A1S4F6D1 A0A182H3F7 D2A3N4 Q17DV6 G3EIW9 A0A336M056 A0A336M2J5 A0A182VDS3 A0A182WV76 A0A182LAT9 A0A182I8V1 Q7PFS6 E2A5J5 A0A2J7QH94 A0A182SY41 A0A182TTM6 F4ZNH7 A0A067QS81 A0A182VXW9 A0A182MAY6 E0VG36 A0A0M9AA53 A0A182R5V2 A0A182K6F1 A0A139WFX6 Q17DV5 A0A154PD41 A0A084VGK6 A0A1J1HR16 A0A182IZU5 A0A0A9VXY2 A0A088AJU4 A0A3L8D5U9 G3EIW8 B4MT95 A0A182H3F8 A0A1I8NUQ1 A0A2A3EHS4 A0A1S4F6C0 A0A158NUU8 G3EIW7 A0A310SX81 B4JKZ1 Q29IT2 A0A195BAK2 K7J0B9 A0A182F7P4 A0A2S2PXF2 A0A182H5D1 A0A195E3M0 A0A195FTT2 A0A1A9XSX2 A0A0L7RJU0 A0A182LZG9 W5JAQ1 B4M6M0 E2BJJ7 B4GEE8 E1ZVM3 A0A182U7N2 A0A182JLA4 A0A3B0JDI1 A0A182HSQ7 F4ZNH5 A0A232EID2 B4QW34 A0A2H1V5E5 A0A182UYY5 Q297A4 F4WAD9 A0A1S4GF89 A0A182LES8 Q7QCC0 A0A0L7L1A4 B4HAM2
A0A2A4JIB2 A0A437BIJ6 A0A212FMP4 A0A0L7LG80 A0A437BXD9 A0A0L7KWS0 A0A2W1BSS3 A0A212EHV5 I4DN78 A0A194QIN4 A0A212FMN5 A0A194RJ52 A0A194PID4 A0A194R0C6 A0A2H1VEA7 H9JTZ7 A0A0L7LTW1 A0A1B0D7W4 A0A1J1IYP0 A0A1S4F6D1 A0A182H3F7 D2A3N4 Q17DV6 G3EIW9 A0A336M056 A0A336M2J5 A0A182VDS3 A0A182WV76 A0A182LAT9 A0A182I8V1 Q7PFS6 E2A5J5 A0A2J7QH94 A0A182SY41 A0A182TTM6 F4ZNH7 A0A067QS81 A0A182VXW9 A0A182MAY6 E0VG36 A0A0M9AA53 A0A182R5V2 A0A182K6F1 A0A139WFX6 Q17DV5 A0A154PD41 A0A084VGK6 A0A1J1HR16 A0A182IZU5 A0A0A9VXY2 A0A088AJU4 A0A3L8D5U9 G3EIW8 B4MT95 A0A182H3F8 A0A1I8NUQ1 A0A2A3EHS4 A0A1S4F6C0 A0A158NUU8 G3EIW7 A0A310SX81 B4JKZ1 Q29IT2 A0A195BAK2 K7J0B9 A0A182F7P4 A0A2S2PXF2 A0A182H5D1 A0A195E3M0 A0A195FTT2 A0A1A9XSX2 A0A0L7RJU0 A0A182LZG9 W5JAQ1 B4M6M0 E2BJJ7 B4GEE8 E1ZVM3 A0A182U7N2 A0A182JLA4 A0A3B0JDI1 A0A182HSQ7 F4ZNH5 A0A232EID2 B4QW34 A0A2H1V5E5 A0A182UYY5 Q297A4 F4WAD9 A0A1S4GF89 A0A182LES8 Q7QCC0 A0A0L7L1A4 B4HAM2
PDB
5NCC
E-value=2.08957e-31,
Score=339
Ontologies
GO
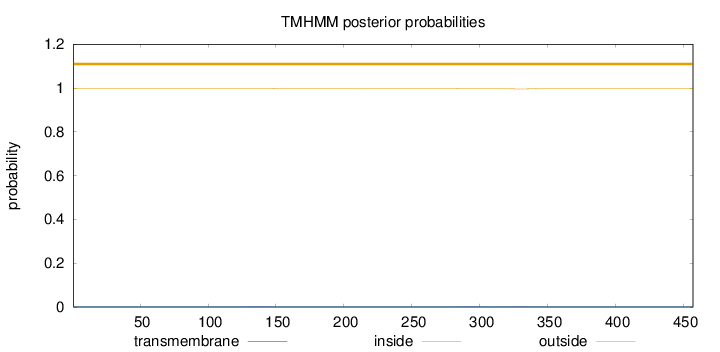
Topology
Length:
457
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0839999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00335
outside
1 - 457
Population Genetic Test Statistics
Pi
188.762321
Theta
207.217883
Tajima's D
-0.693465
CLR
1.286834
CSRT
0.199890005499725
Interpretation
Uncertain
