Gene
KWMTBOMO09571
Pre Gene Modal
BGIBMGA012834
Annotation
PREDICTED:_NAD-dependent_protein_deacylase_Sirt4_[Amyelois_transitella]
Full name
NAD-dependent protein deacylase
+ More
NAD-dependent protein deacylase Sirt4
NAD-dependent protein deacylase Sirt4
Alternative Name
Regulatory protein SIR2 homolog
Regulatory protein SIR2 homolog 4
Regulatory protein SIR2 homolog 4
Location in the cell
Nuclear Reliability : 2.792
Sequence
CDS
ATGTTAAAAAGATTTATACCGTTTAATAATAACGCTATTAGCAGACACATAGTGTGTGTTCCGAAACATAAACCACCCGAAAAAAAAGATTTTGACGTTTTGAAAGAGTTCTTAACAAACCATAACAAAATTTTAATTTTGACTGGTGCTGGCATTTCTACTGAATCAGGTATTCCTGACTACAGATCGGAGGATGTTGGTCTGTATGCCCGTAGTAATCACAAACCTATACAGTATCAAGAATTTGTGAAGTATCCGAAAGTAAGGCAAAGGTACTGGGCAAGAAATTATATAGGTTGGCCGAGATTCAGTTGTGTGCAACCGAACGTCACACATCTCTGTATTCGAGAACTAGAGAAGAAGGGAAAAGTGACATCTATAGTGACACAGAATGTGGATAGATTACACCACAAAGCTGGTTCTGAAAAAGTGATTGAATTACATGGGACAAGCTACTTAGTTCAATGTCTCAAATGCCCCTATGAAATAGACAGACATGAACTCCAAGAAATATTAACAGAGAATAATCCAGATATGGAAAGTAGTTTCTCCATGATAAGACCGGATGGCGATGTTGAATTGTCGAGGGAACAAGTAGAAAAATTCCGAGCTCCACTCTGCCCAAAGTGTGAAGGGCCTTTGAAACCAGATATAGTGTTTTTTGGTGACAATGTACCCAAATATCGCGTGGAACAAGTCAGAAAAGAAGTGACTAGCAGTGATGCTGTATTTGTAATGGGATCAAGCCTCACAGTTTATTCTAGCTATAGAATTATATTGCAAGCCAGAGATGAACACAAAAACATAGCTATATTAAATATTGGACCTACTAGAGCAGATGATATTGTTAATATAAAAGTATCTACAAAATGTGGTGATGTTTTACCTGAATTATGTAAATCTTTAAATTAA
Protein
MLKRFIPFNNNAISRHIVCVPKHKPPEKKDFDVLKEFLTNHNKILILTGAGISTESGIPDYRSEDVGLYARSNHKPIQYQEFVKYPKVRQRYWARNYIGWPRFSCVQPNVTHLCIRELEKKGKVTSIVTQNVDRLHHKAGSEKVIELHGTSYLVQCLKCPYEIDRHELQEILTENNPDMESSFSMIRPDGDVELSREQVEKFRAPLCPKCEGPLKPDIVFFGDNVPKYRVEQVRKEVTSSDAVFVMGSSLTVYSSYRIILQARDEHKNIAILNIGPTRADDIVNIKVSTKCGDVLPELCKSLN
Summary
Description
NAD-dependent protein deacylase. Catalyzes the NAD-dependent hydrolysis of acyl groups from lysine residues.
Catalytic Activity
H2O + N(6)-acetyl-L-lysyl-[protein] + NAD(+) = 2''-O-acetyl-ADP-D-ribose + L-lysyl-[protein] + nicotinamide
Cofactor
Zn(2+)
Similarity
Belongs to the sirtuin family. Class II subfamily.
Keywords
Alternative splicing
Complete proteome
Hydrolase
Metal-binding
Mitochondrion
NAD
Reference proteome
Transit peptide
Zinc
Feature
chain NAD-dependent protein deacylase Sirt4
splice variant In isoform a.
splice variant In isoform a.
Uniprot
H9JTH2
A0A2H1VEA1
A0A2A4J2K7
A0A194RDQ5
A0A194QIB3
A0A0L7L9D9
+ More
A0A3S2M3T3 A0A212FMM6 A0A139WG94 A0A2J7PRA2 A0A1S4ELI4 E2ADS2 E9J587 A0A1Y1M4Y1 A0A0T6B3I2 A0A0K8UKC0 A0A1Y1M0E0 A0A067R0A4 A0A2S2NA90 A0A182JVE2 A0A182XXF5 A0A034WQI7 A0A182P7A2 A0A182H5D4 A0A195EUE3 A0A1B6H4G9 E2BN27 A0A182H3G2 A0A1I8MSC8 A0A182I8V4 A0A182WV79 A0A182J7Z3 F5HJ98 B0W3I7 A0A182R5U9 A0A158NR58 A0A3F2Z106 A0A182QZ02 A0A182SLH5 A0A182UTK3 A0A2A3EB70 A0A182N4G3 A0A1B6KFT4 A0A182MJ25 A0A1B6K1X5 A0A182F7P2 A0A1Q3F775 Q17DV2 W5JBK9 A0A1S4F6N7 A0A310SR36 A0A195E5T4 A0A2M4ATK5 A0A195B0X7 J9K8C7 F4WPE5 A0A336LMW1 A0A151I6S8 A0A1I8PPI2 A0A423TP43 A0A088A4P0 A0A151WMA8 B3MRX4 T1JHL2 A0A1A9XC19 A0A1W4VF60 B3NV66 B4L644 A0A1A9VB17 B4R505 A0A0C9PWP9 B4PZD0 W8ANG2 Q8IRR5 B4I0M1 B4M804 A0A1W4XD59 C4WX51 A0A0M3QZM2 A0A0M8ZU10 A0A026WFF3 A0A0P5PRC6 A0A1B6BZS2 B4MSG3 A0A0P5ATX6 E9G957 A0A0P5II33 T1PB20 K7IS10 A0A1W4X3M4 N6TQF1 A0A0K8TT56 B4JKI2 A0A232EES1 K1R299 A0A0P5MKR8 A0A1Y1M5X7 Q29I14 B4H4G5 A0A2C9JZL2
A0A3S2M3T3 A0A212FMM6 A0A139WG94 A0A2J7PRA2 A0A1S4ELI4 E2ADS2 E9J587 A0A1Y1M4Y1 A0A0T6B3I2 A0A0K8UKC0 A0A1Y1M0E0 A0A067R0A4 A0A2S2NA90 A0A182JVE2 A0A182XXF5 A0A034WQI7 A0A182P7A2 A0A182H5D4 A0A195EUE3 A0A1B6H4G9 E2BN27 A0A182H3G2 A0A1I8MSC8 A0A182I8V4 A0A182WV79 A0A182J7Z3 F5HJ98 B0W3I7 A0A182R5U9 A0A158NR58 A0A3F2Z106 A0A182QZ02 A0A182SLH5 A0A182UTK3 A0A2A3EB70 A0A182N4G3 A0A1B6KFT4 A0A182MJ25 A0A1B6K1X5 A0A182F7P2 A0A1Q3F775 Q17DV2 W5JBK9 A0A1S4F6N7 A0A310SR36 A0A195E5T4 A0A2M4ATK5 A0A195B0X7 J9K8C7 F4WPE5 A0A336LMW1 A0A151I6S8 A0A1I8PPI2 A0A423TP43 A0A088A4P0 A0A151WMA8 B3MRX4 T1JHL2 A0A1A9XC19 A0A1W4VF60 B3NV66 B4L644 A0A1A9VB17 B4R505 A0A0C9PWP9 B4PZD0 W8ANG2 Q8IRR5 B4I0M1 B4M804 A0A1W4XD59 C4WX51 A0A0M3QZM2 A0A0M8ZU10 A0A026WFF3 A0A0P5PRC6 A0A1B6BZS2 B4MSG3 A0A0P5ATX6 E9G957 A0A0P5II33 T1PB20 K7IS10 A0A1W4X3M4 N6TQF1 A0A0K8TT56 B4JKI2 A0A232EES1 K1R299 A0A0P5MKR8 A0A1Y1M5X7 Q29I14 B4H4G5 A0A2C9JZL2
EC Number
3.5.1.-
Pubmed
19121390
26354079
26227816
22118469
18362917
19820115
+ More
20798317 21282665 28004739 24845553 25244985 25348373 26483478 25315136 12364791 14747013 17210077 21347285 17510324 20920257 23761445 21719571 17994087 17550304 24495485 10731132 12537572 24508170 21292972 20075255 23537049 26369729 28648823 22992520 15632085 15562597
20798317 21282665 28004739 24845553 25244985 25348373 26483478 25315136 12364791 14747013 17210077 21347285 17510324 20920257 23761445 21719571 17994087 17550304 24495485 10731132 12537572 24508170 21292972 20075255 23537049 26369729 28648823 22992520 15632085 15562597
EMBL
BABH01003834
ODYU01002091
SOQ39163.1
NWSH01003390
PCG66387.1
KQ460323
+ More
KPJ15973.1 KQ458756 KPJ05257.1 JTDY01002148 KOB72000.1 RSAL01000051 RVE50273.1 AGBW02007651 OWR55014.1 KQ971348 KYB26875.1 NEVH01022362 PNF18868.1 GL438820 EFN68434.1 GL768124 EFZ12017.1 GEZM01043124 GEZM01043121 GEZM01043114 GEZM01043113 JAV79325.1 LJIG01016066 KRT81717.1 GDHF01025180 JAI27134.1 GEZM01043119 JAV79312.1 KK852802 KDR16289.1 GGMR01001253 MBY13872.1 GAKP01002365 JAC56587.1 JXUM01027346 KQ560787 KXJ80886.1 KQ981965 KYN31868.1 GECZ01000219 JAS69550.1 GL449382 EFN82931.1 JXUM01024572 KQ560694 KXJ81246.1 APCN01003125 AAAB01008844 EGK96359.1 DS231832 EDS31773.1 ADTU01023713 AXCN02000040 KZ288295 PBC28968.1 GEBQ01029684 JAT10293.1 AXCM01002816 GECU01002265 JAT05442.1 GFDL01011625 JAV23420.1 CH477290 EAT44649.1 ADMH02001961 ETN60235.1 KQ760801 OAD58981.1 KQ979608 KYN20446.1 GGFK01010805 MBW44126.1 KQ976691 KYM77945.1 ABLF02021374 GL888243 EGI64040.1 UFQS01000065 UFQT01000065 SSW98920.1 SSX19306.1 KQ978459 KYM93810.1 QCYY01001410 ROT78235.1 KQ982944 KYQ48986.1 CH902622 EDV34529.1 JH431506 CH954180 EDV45914.1 CH933812 EDW05840.1 CM000366 EDX17154.1 GBYB01005933 JAG75700.1 CM000162 EDX01097.1 GAMC01016215 JAB90340.1 AE014298 BT011040 BT099555 BT100157 CH480819 EDW53052.1 CH940653 EDW62280.1 AK342451 BAH72471.1 CP012528 ALC49663.1 KQ435885 KOX69603.1 KK107273 EZA53754.1 GDIQ01130949 GDIQ01089516 JAL20777.1 GEDC01030535 JAS06763.1 CH963851 EDW75052.1 GDIP01194148 JAJ29254.1 GL732535 EFX84150.1 GDIQ01214469 JAK37256.1 KA645951 AFP60580.1 APGK01055725 APGK01055726 APGK01055727 KB741269 ENN71465.1 GDAI01000272 JAI17331.1 CH916370 EDW00085.1 NNAY01005248 OXU16828.1 JH817068 EKC35255.1 GDIQ01155137 JAK96588.1 GEZM01043122 GEZM01043118 JAV79306.1 CH379064 EAL31592.1 CH479209 EDW32716.1
KPJ15973.1 KQ458756 KPJ05257.1 JTDY01002148 KOB72000.1 RSAL01000051 RVE50273.1 AGBW02007651 OWR55014.1 KQ971348 KYB26875.1 NEVH01022362 PNF18868.1 GL438820 EFN68434.1 GL768124 EFZ12017.1 GEZM01043124 GEZM01043121 GEZM01043114 GEZM01043113 JAV79325.1 LJIG01016066 KRT81717.1 GDHF01025180 JAI27134.1 GEZM01043119 JAV79312.1 KK852802 KDR16289.1 GGMR01001253 MBY13872.1 GAKP01002365 JAC56587.1 JXUM01027346 KQ560787 KXJ80886.1 KQ981965 KYN31868.1 GECZ01000219 JAS69550.1 GL449382 EFN82931.1 JXUM01024572 KQ560694 KXJ81246.1 APCN01003125 AAAB01008844 EGK96359.1 DS231832 EDS31773.1 ADTU01023713 AXCN02000040 KZ288295 PBC28968.1 GEBQ01029684 JAT10293.1 AXCM01002816 GECU01002265 JAT05442.1 GFDL01011625 JAV23420.1 CH477290 EAT44649.1 ADMH02001961 ETN60235.1 KQ760801 OAD58981.1 KQ979608 KYN20446.1 GGFK01010805 MBW44126.1 KQ976691 KYM77945.1 ABLF02021374 GL888243 EGI64040.1 UFQS01000065 UFQT01000065 SSW98920.1 SSX19306.1 KQ978459 KYM93810.1 QCYY01001410 ROT78235.1 KQ982944 KYQ48986.1 CH902622 EDV34529.1 JH431506 CH954180 EDV45914.1 CH933812 EDW05840.1 CM000366 EDX17154.1 GBYB01005933 JAG75700.1 CM000162 EDX01097.1 GAMC01016215 JAB90340.1 AE014298 BT011040 BT099555 BT100157 CH480819 EDW53052.1 CH940653 EDW62280.1 AK342451 BAH72471.1 CP012528 ALC49663.1 KQ435885 KOX69603.1 KK107273 EZA53754.1 GDIQ01130949 GDIQ01089516 JAL20777.1 GEDC01030535 JAS06763.1 CH963851 EDW75052.1 GDIP01194148 JAJ29254.1 GL732535 EFX84150.1 GDIQ01214469 JAK37256.1 KA645951 AFP60580.1 APGK01055725 APGK01055726 APGK01055727 KB741269 ENN71465.1 GDAI01000272 JAI17331.1 CH916370 EDW00085.1 NNAY01005248 OXU16828.1 JH817068 EKC35255.1 GDIQ01155137 JAK96588.1 GEZM01043122 GEZM01043118 JAV79306.1 CH379064 EAL31592.1 CH479209 EDW32716.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000037510
UP000283053
+ More
UP000007151 UP000007266 UP000235965 UP000079169 UP000000311 UP000027135 UP000075881 UP000076408 UP000075885 UP000069940 UP000249989 UP000078541 UP000008237 UP000095301 UP000075840 UP000076407 UP000075880 UP000007062 UP000002320 UP000075900 UP000005205 UP000075920 UP000075886 UP000075901 UP000075903 UP000242457 UP000075884 UP000075883 UP000069272 UP000008820 UP000000673 UP000078492 UP000078540 UP000007819 UP000007755 UP000078542 UP000095300 UP000283509 UP000005203 UP000075809 UP000007801 UP000092443 UP000192221 UP000008711 UP000009192 UP000078200 UP000000304 UP000002282 UP000000803 UP000001292 UP000008792 UP000192223 UP000092553 UP000053105 UP000053097 UP000007798 UP000000305 UP000002358 UP000019118 UP000001070 UP000215335 UP000005408 UP000001819 UP000008744 UP000076420
UP000007151 UP000007266 UP000235965 UP000079169 UP000000311 UP000027135 UP000075881 UP000076408 UP000075885 UP000069940 UP000249989 UP000078541 UP000008237 UP000095301 UP000075840 UP000076407 UP000075880 UP000007062 UP000002320 UP000075900 UP000005205 UP000075920 UP000075886 UP000075901 UP000075903 UP000242457 UP000075884 UP000075883 UP000069272 UP000008820 UP000000673 UP000078492 UP000078540 UP000007819 UP000007755 UP000078542 UP000095300 UP000283509 UP000005203 UP000075809 UP000007801 UP000092443 UP000192221 UP000008711 UP000009192 UP000078200 UP000000304 UP000002282 UP000000803 UP000001292 UP000008792 UP000192223 UP000092553 UP000053105 UP000053097 UP000007798 UP000000305 UP000002358 UP000019118 UP000001070 UP000215335 UP000005408 UP000001819 UP000008744 UP000076420
Interpro
IPR029035
DHS-like_NAD/FAD-binding_dom
+ More
IPR026591 Sirtuin_cat_small_dom_sf
IPR003000 Sirtuin
IPR026590 Ssirtuin_cat_dom
IPR026587 Sirtuin_class_II
IPR006612 THAP_Znf
IPR038441 THAP_Znf_sf
IPR009057 Homeobox-like_sf
IPR007889 HTH_Psq
IPR006600 HTH_CenpB_DNA-bd_dom
IPR004875 DDE_SF_endonuclease_dom
IPR026591 Sirtuin_cat_small_dom_sf
IPR003000 Sirtuin
IPR026590 Ssirtuin_cat_dom
IPR026587 Sirtuin_class_II
IPR006612 THAP_Znf
IPR038441 THAP_Znf_sf
IPR009057 Homeobox-like_sf
IPR007889 HTH_Psq
IPR006600 HTH_CenpB_DNA-bd_dom
IPR004875 DDE_SF_endonuclease_dom
Gene 3D
ProteinModelPortal
H9JTH2
A0A2H1VEA1
A0A2A4J2K7
A0A194RDQ5
A0A194QIB3
A0A0L7L9D9
+ More
A0A3S2M3T3 A0A212FMM6 A0A139WG94 A0A2J7PRA2 A0A1S4ELI4 E2ADS2 E9J587 A0A1Y1M4Y1 A0A0T6B3I2 A0A0K8UKC0 A0A1Y1M0E0 A0A067R0A4 A0A2S2NA90 A0A182JVE2 A0A182XXF5 A0A034WQI7 A0A182P7A2 A0A182H5D4 A0A195EUE3 A0A1B6H4G9 E2BN27 A0A182H3G2 A0A1I8MSC8 A0A182I8V4 A0A182WV79 A0A182J7Z3 F5HJ98 B0W3I7 A0A182R5U9 A0A158NR58 A0A3F2Z106 A0A182QZ02 A0A182SLH5 A0A182UTK3 A0A2A3EB70 A0A182N4G3 A0A1B6KFT4 A0A182MJ25 A0A1B6K1X5 A0A182F7P2 A0A1Q3F775 Q17DV2 W5JBK9 A0A1S4F6N7 A0A310SR36 A0A195E5T4 A0A2M4ATK5 A0A195B0X7 J9K8C7 F4WPE5 A0A336LMW1 A0A151I6S8 A0A1I8PPI2 A0A423TP43 A0A088A4P0 A0A151WMA8 B3MRX4 T1JHL2 A0A1A9XC19 A0A1W4VF60 B3NV66 B4L644 A0A1A9VB17 B4R505 A0A0C9PWP9 B4PZD0 W8ANG2 Q8IRR5 B4I0M1 B4M804 A0A1W4XD59 C4WX51 A0A0M3QZM2 A0A0M8ZU10 A0A026WFF3 A0A0P5PRC6 A0A1B6BZS2 B4MSG3 A0A0P5ATX6 E9G957 A0A0P5II33 T1PB20 K7IS10 A0A1W4X3M4 N6TQF1 A0A0K8TT56 B4JKI2 A0A232EES1 K1R299 A0A0P5MKR8 A0A1Y1M5X7 Q29I14 B4H4G5 A0A2C9JZL2
A0A3S2M3T3 A0A212FMM6 A0A139WG94 A0A2J7PRA2 A0A1S4ELI4 E2ADS2 E9J587 A0A1Y1M4Y1 A0A0T6B3I2 A0A0K8UKC0 A0A1Y1M0E0 A0A067R0A4 A0A2S2NA90 A0A182JVE2 A0A182XXF5 A0A034WQI7 A0A182P7A2 A0A182H5D4 A0A195EUE3 A0A1B6H4G9 E2BN27 A0A182H3G2 A0A1I8MSC8 A0A182I8V4 A0A182WV79 A0A182J7Z3 F5HJ98 B0W3I7 A0A182R5U9 A0A158NR58 A0A3F2Z106 A0A182QZ02 A0A182SLH5 A0A182UTK3 A0A2A3EB70 A0A182N4G3 A0A1B6KFT4 A0A182MJ25 A0A1B6K1X5 A0A182F7P2 A0A1Q3F775 Q17DV2 W5JBK9 A0A1S4F6N7 A0A310SR36 A0A195E5T4 A0A2M4ATK5 A0A195B0X7 J9K8C7 F4WPE5 A0A336LMW1 A0A151I6S8 A0A1I8PPI2 A0A423TP43 A0A088A4P0 A0A151WMA8 B3MRX4 T1JHL2 A0A1A9XC19 A0A1W4VF60 B3NV66 B4L644 A0A1A9VB17 B4R505 A0A0C9PWP9 B4PZD0 W8ANG2 Q8IRR5 B4I0M1 B4M804 A0A1W4XD59 C4WX51 A0A0M3QZM2 A0A0M8ZU10 A0A026WFF3 A0A0P5PRC6 A0A1B6BZS2 B4MSG3 A0A0P5ATX6 E9G957 A0A0P5II33 T1PB20 K7IS10 A0A1W4X3M4 N6TQF1 A0A0K8TT56 B4JKI2 A0A232EES1 K1R299 A0A0P5MKR8 A0A1Y1M5X7 Q29I14 B4H4G5 A0A2C9JZL2
PDB
5OJN
E-value=3.26209e-78,
Score=741
Ontologies
GO
Topology
Subcellular location
Mitochondrion matrix
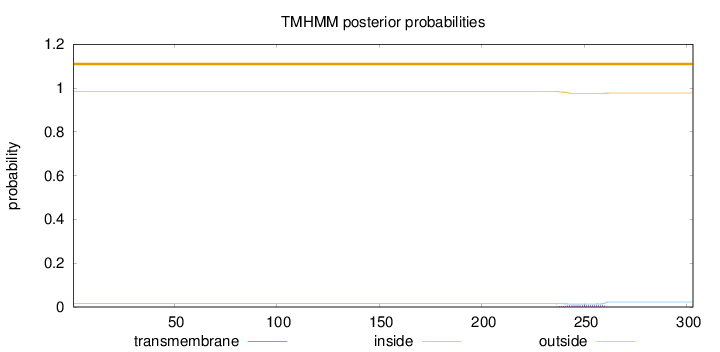
Length:
303
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.20358
Exp number, first 60 AAs:
0.00098
Total prob of N-in:
0.01471
outside
1 - 303
Population Genetic Test Statistics
Pi
20.231867
Theta
17.77057
Tajima's D
0.218922
CLR
1.516999
CSRT
0.44302784860757
Interpretation
Uncertain
