Gene
KWMTBOMO09568
Pre Gene Modal
BGIBMGA013012
Annotation
olfactory_receptor_2_[Bombyx_mori]
Full name
Odorant receptor
Location in the cell
PlasmaMembrane Reliability : 3.052
Sequence
CDS
ATGATGACCAAGGTCAAGACGCAGGGCCTGGTCACAGACCTGATGCCCTGCATAAGATTACTGCAAGCAGCGGGCCACTTCTTATTCAATTATCATGCCGATACCTCCGGAATGAATATGCTCCTACGGAAAATATATTCGAGTGCCCATGCCGTGCTCATTGTCGTCCACTACATCTGCATGGGCATCAACATGGCACAATACAAGGACGAAGTCAACGAGCTCACGGCAAATACCATTACGGTCCTGTTTTTCGCTCACAGCATCATTAAGTTGGCATTCTTTGCGTTCAACTCTAAAAGTTTTTACAGGTAA
Protein
MMTKVKTQGLVTDLMPCIRLLQAAGHFLFNYHADTSGMNMLLRKIYSSAHAVLIVVHYICMGINMAQYKDEVNELTANTITVLFFAHSIIKLAFFAFNSKSFYR
Summary
Similarity
Belongs to the insect chemoreceptor superfamily. Heteromeric odorant receptor channel (TC 1.A.69) family.
Uniprot
Q7YT34
H9JU00
A0A0S1U2I3
B5MEJ8
I3Y3I7
A0A0U2RFK0
+ More
H9XFD5 A7UGE1 A0A1B3B711 B5MEJ5 B6C755 A0A146JVN3 A0A1U9W0P1 Q8MMI4 A0A0B5A1E6 A0A075TD45 I1V1V9 E5KJL4 E5KJL5 A0A2H1VPE2 Q0MUT5 Q5I2A3 B6VCU1 A0A0P0F2D8 R9QVV2 U5NFY8 Q5D7J6 Q5QHV7 J7KF99 C1PHA7 M4NL03 A0A0E4B591 A0A1S5XXL1 A0A0P1JD94 A0A223HDF7 E7E207 A0A3G6V705 J7G1V0 D3J5H5 A0A223HD99 Q7YT35 A0A2R4SBD4 G9BLV3 A0A2K8GKR9 A0A0B5CN11 G9BLV6 G9BLV5 A0A0B5GID1 A0A097ITX9 G9BLV4 G9BLV2 A0A097IYL2 U5KJY5 D0EZE1 A0A194R291 A0A223HD43 C4MX28 B5MEK1 H9A5M2 A0A076E9A8 C1PHA8 E2S067 A0A076E5W5 E2S069 A0A1B3P5R0 A0A386NB18 A0A386H9D4 A0A0K0KG02 E2S068 E2S071 E2S072 F5CJE4 A0A345BET1 A0A212EN65 A0A140YWR4 E2S070 A0A0L7LQN7 A0A220D5Z4 A0A3S7SGT1 A0A0K1W013 A0A2H4NT96 A0A2Z4EY25 B2ZDZ6 B9VH08 A0A3G2KX47 A0A2P1JH84 S5U415 A0A0B5ITU4 A0A0L7QV02 A0A0U4DIR1 E0VHN8 A0A0H3W578 A0A0N9JVJ7 C0Z3R3 A0A0S3J2L6 A0A0C9PWC1 A0A1S5VFJ4 V9MVU4 G9J1I3 A1YWY0
H9XFD5 A7UGE1 A0A1B3B711 B5MEJ5 B6C755 A0A146JVN3 A0A1U9W0P1 Q8MMI4 A0A0B5A1E6 A0A075TD45 I1V1V9 E5KJL4 E5KJL5 A0A2H1VPE2 Q0MUT5 Q5I2A3 B6VCU1 A0A0P0F2D8 R9QVV2 U5NFY8 Q5D7J6 Q5QHV7 J7KF99 C1PHA7 M4NL03 A0A0E4B591 A0A1S5XXL1 A0A0P1JD94 A0A223HDF7 E7E207 A0A3G6V705 J7G1V0 D3J5H5 A0A223HD99 Q7YT35 A0A2R4SBD4 G9BLV3 A0A2K8GKR9 A0A0B5CN11 G9BLV6 G9BLV5 A0A0B5GID1 A0A097ITX9 G9BLV4 G9BLV2 A0A097IYL2 U5KJY5 D0EZE1 A0A194R291 A0A223HD43 C4MX28 B5MEK1 H9A5M2 A0A076E9A8 C1PHA8 E2S067 A0A076E5W5 E2S069 A0A1B3P5R0 A0A386NB18 A0A386H9D4 A0A0K0KG02 E2S068 E2S071 E2S072 F5CJE4 A0A345BET1 A0A212EN65 A0A140YWR4 E2S070 A0A0L7LQN7 A0A220D5Z4 A0A3S7SGT1 A0A0K1W013 A0A2H4NT96 A0A2Z4EY25 B2ZDZ6 B9VH08 A0A3G2KX47 A0A2P1JH84 S5U415 A0A0B5ITU4 A0A0L7QV02 A0A0U4DIR1 E0VHN8 A0A0H3W578 A0A0N9JVJ7 C0Z3R3 A0A0S3J2L6 A0A0C9PWC1 A0A1S5VFJ4 V9MVU4 G9J1I3 A1YWY0
Pubmed
12827420
15545611
19121390
22646846
26657286
18057325
+ More
18828844 12270037 25027790 28756777 23195879 26812239 23894529 25039606 15723778 23950722 28150741 30465940 22911835 20084285 22701634 28736530 25252791 24130875 26354079 19293399 22363688 27006164 24998398 27538507 29727827 22118469 26729427 26227816 29126322 29894821 30319455 29497384 23964849 26494014 20566863 25938508 26164593 18362917 19820115 26626891 28187311
18828844 12270037 25027790 28756777 23195879 26812239 23894529 25039606 15723778 23950722 28150741 30465940 22911835 20084285 22701634 28736530 25252791 24130875 26354079 19293399 22363688 27006164 24998398 27538507 29727827 22118469 26729427 26227816 29126322 29894821 30319455 29497384 23964849 26494014 20566863 25938508 26164593 18362917 19820115 26626891 28187311
EMBL
AB100454
AJ555487
BAD69585.1
CAD88206.1
BABH01003841
KT020861
+ More
ALM30348.1 AB263114 BAG71418.1 JQ794817 AFL70822.1 KP975160 ALT31679.1 JN867062 AFG73001.1 EU057178 ABU45983.2 KT588097 AOE48007.1 AB263111 GENK01000117 BAG71415.1 JAV45796.1 EF395366 ABQ82137.1 KX084453 GEDO01000046 ARO76408.1 JAP88580.1 KY365529 AQY16483.1 AJ487477 NWSH01000897 CAD31851.1 PCG73648.1 KJ542658 AJD81542.1 KF768657 KZ150073 AIG51851.1 PZC74007.1 JQ394904 AFI25169.1 HQ186284 ADQ13177.1 HQ186285 ADQ13178.1 ODYU01003404 SOQ42114.1 DQ845292 JQ811935 ABH10019.1 AFN22085.1 AY862142 AAW52583.1 FJ374688 ACJ06648.1 KR632987 ALJ33155.1 JX101681 AGF29886.1 KC960454 AGY14565.1 AY843204 AAX14773.1 AY485222 AAS49925.1 JX390608 RSAL01000093 AFQ94048.1 RVE47931.1 AB467318 BAH57973.1 JX910526 AGG91643.1 LC002697 BAR43445.1 KY399256 AQQ73487.1 LN885133 CUQ99422.1 KY283761 AST36420.1 HQ596409 ADT82677.1 MH193973 AZB49419.1 JX173647 AFP54145.1 GQ844877 ADB89179.1 KY283682 AST36341.1 AJ555486 CAD88205.1 MG581972 AVZ44715.1 HQ619212 AET06156.1 KX585321 ARO70214.1 KM678337 AJE25910.1 HQ619215 AET06159.1 HQ619214 AET06158.1 KM892411 AJF23826.1 KM597236 AIT69913.1 HQ619213 AET06157.1 HQ619211 AET06155.1 KM655573 AIT72022.1 KC526964 AGS41440.1 GQ923610 ACX54944.1 KQ460878 KPJ11634.1 KY283633 AST36292.1 EU791887 ACJ12928.2 AB263117 BAG71421.2 JN836672 AFC91712.1 KF487681 AII01079.1 AB467319 BAH57974.1 AB595945 BAJ23260.1 KF487648 AII01046.1 AB595947 BAJ23262.1 KX655981 AOG12930.1 MG587081 AYE19959.1 MG546597 AYD42220.1 KF513149 AII15784.1 AB595946 BAJ23261.1 AB595949 BAJ23264.1 AB595950 BAJ23265.1 JF714981 AEA76288.1 MG816570 AXF48755.1 AGBW02013713 OWR42934.1 KJ592712 AJF20962.1 AB595948 BAJ23263.1 JTDY01000319 KOB77714.1 KX618692 ARQ32245.1 MF625584 AXY83411.1 KT373968 AKW50880.1 KY750554 ATV96621.1 MG913127 AWV67916.1 EU647221 ACD40044.1 FJ546087 ACM18060.1 MH723588 AYN64391.1 MG766143 AVN97813.1 KC778527 KB465818 AGS43074.1 RLZ02183.1 KM893996 AJF94638.2 KQ414731 KOC62379.1 KT369093 ALX17413.1 DS235171 EEB12924.1 KP736145 AKK25151.1 KP963696 ALG36144.1 AM689918 KQ971345 CAM84014.1 EFA05687.1 KT381541 ALR72547.1 GBYB01005773 GBYB01005774 GBYB01013276 JAG75540.1 JAG75541.1 JAG83043.1 KY445468 AQN78403.1 JX472452 AGI62937.2 JN792581 AET85154.2 EF141511 ABM05966.1
ALM30348.1 AB263114 BAG71418.1 JQ794817 AFL70822.1 KP975160 ALT31679.1 JN867062 AFG73001.1 EU057178 ABU45983.2 KT588097 AOE48007.1 AB263111 GENK01000117 BAG71415.1 JAV45796.1 EF395366 ABQ82137.1 KX084453 GEDO01000046 ARO76408.1 JAP88580.1 KY365529 AQY16483.1 AJ487477 NWSH01000897 CAD31851.1 PCG73648.1 KJ542658 AJD81542.1 KF768657 KZ150073 AIG51851.1 PZC74007.1 JQ394904 AFI25169.1 HQ186284 ADQ13177.1 HQ186285 ADQ13178.1 ODYU01003404 SOQ42114.1 DQ845292 JQ811935 ABH10019.1 AFN22085.1 AY862142 AAW52583.1 FJ374688 ACJ06648.1 KR632987 ALJ33155.1 JX101681 AGF29886.1 KC960454 AGY14565.1 AY843204 AAX14773.1 AY485222 AAS49925.1 JX390608 RSAL01000093 AFQ94048.1 RVE47931.1 AB467318 BAH57973.1 JX910526 AGG91643.1 LC002697 BAR43445.1 KY399256 AQQ73487.1 LN885133 CUQ99422.1 KY283761 AST36420.1 HQ596409 ADT82677.1 MH193973 AZB49419.1 JX173647 AFP54145.1 GQ844877 ADB89179.1 KY283682 AST36341.1 AJ555486 CAD88205.1 MG581972 AVZ44715.1 HQ619212 AET06156.1 KX585321 ARO70214.1 KM678337 AJE25910.1 HQ619215 AET06159.1 HQ619214 AET06158.1 KM892411 AJF23826.1 KM597236 AIT69913.1 HQ619213 AET06157.1 HQ619211 AET06155.1 KM655573 AIT72022.1 KC526964 AGS41440.1 GQ923610 ACX54944.1 KQ460878 KPJ11634.1 KY283633 AST36292.1 EU791887 ACJ12928.2 AB263117 BAG71421.2 JN836672 AFC91712.1 KF487681 AII01079.1 AB467319 BAH57974.1 AB595945 BAJ23260.1 KF487648 AII01046.1 AB595947 BAJ23262.1 KX655981 AOG12930.1 MG587081 AYE19959.1 MG546597 AYD42220.1 KF513149 AII15784.1 AB595946 BAJ23261.1 AB595949 BAJ23264.1 AB595950 BAJ23265.1 JF714981 AEA76288.1 MG816570 AXF48755.1 AGBW02013713 OWR42934.1 KJ592712 AJF20962.1 AB595948 BAJ23263.1 JTDY01000319 KOB77714.1 KX618692 ARQ32245.1 MF625584 AXY83411.1 KT373968 AKW50880.1 KY750554 ATV96621.1 MG913127 AWV67916.1 EU647221 ACD40044.1 FJ546087 ACM18060.1 MH723588 AYN64391.1 MG766143 AVN97813.1 KC778527 KB465818 AGS43074.1 RLZ02183.1 KM893996 AJF94638.2 KQ414731 KOC62379.1 KT369093 ALX17413.1 DS235171 EEB12924.1 KP736145 AKK25151.1 KP963696 ALG36144.1 AM689918 KQ971345 CAM84014.1 EFA05687.1 KT381541 ALR72547.1 GBYB01005773 GBYB01005774 GBYB01013276 JAG75540.1 JAG75541.1 JAG83043.1 KY445468 AQN78403.1 JX472452 AGI62937.2 JN792581 AET85154.2 EF141511 ABM05966.1
Proteomes
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
Q7YT34
H9JU00
A0A0S1U2I3
B5MEJ8
I3Y3I7
A0A0U2RFK0
+ More
H9XFD5 A7UGE1 A0A1B3B711 B5MEJ5 B6C755 A0A146JVN3 A0A1U9W0P1 Q8MMI4 A0A0B5A1E6 A0A075TD45 I1V1V9 E5KJL4 E5KJL5 A0A2H1VPE2 Q0MUT5 Q5I2A3 B6VCU1 A0A0P0F2D8 R9QVV2 U5NFY8 Q5D7J6 Q5QHV7 J7KF99 C1PHA7 M4NL03 A0A0E4B591 A0A1S5XXL1 A0A0P1JD94 A0A223HDF7 E7E207 A0A3G6V705 J7G1V0 D3J5H5 A0A223HD99 Q7YT35 A0A2R4SBD4 G9BLV3 A0A2K8GKR9 A0A0B5CN11 G9BLV6 G9BLV5 A0A0B5GID1 A0A097ITX9 G9BLV4 G9BLV2 A0A097IYL2 U5KJY5 D0EZE1 A0A194R291 A0A223HD43 C4MX28 B5MEK1 H9A5M2 A0A076E9A8 C1PHA8 E2S067 A0A076E5W5 E2S069 A0A1B3P5R0 A0A386NB18 A0A386H9D4 A0A0K0KG02 E2S068 E2S071 E2S072 F5CJE4 A0A345BET1 A0A212EN65 A0A140YWR4 E2S070 A0A0L7LQN7 A0A220D5Z4 A0A3S7SGT1 A0A0K1W013 A0A2H4NT96 A0A2Z4EY25 B2ZDZ6 B9VH08 A0A3G2KX47 A0A2P1JH84 S5U415 A0A0B5ITU4 A0A0L7QV02 A0A0U4DIR1 E0VHN8 A0A0H3W578 A0A0N9JVJ7 C0Z3R3 A0A0S3J2L6 A0A0C9PWC1 A0A1S5VFJ4 V9MVU4 G9J1I3 A1YWY0
H9XFD5 A7UGE1 A0A1B3B711 B5MEJ5 B6C755 A0A146JVN3 A0A1U9W0P1 Q8MMI4 A0A0B5A1E6 A0A075TD45 I1V1V9 E5KJL4 E5KJL5 A0A2H1VPE2 Q0MUT5 Q5I2A3 B6VCU1 A0A0P0F2D8 R9QVV2 U5NFY8 Q5D7J6 Q5QHV7 J7KF99 C1PHA7 M4NL03 A0A0E4B591 A0A1S5XXL1 A0A0P1JD94 A0A223HDF7 E7E207 A0A3G6V705 J7G1V0 D3J5H5 A0A223HD99 Q7YT35 A0A2R4SBD4 G9BLV3 A0A2K8GKR9 A0A0B5CN11 G9BLV6 G9BLV5 A0A0B5GID1 A0A097ITX9 G9BLV4 G9BLV2 A0A097IYL2 U5KJY5 D0EZE1 A0A194R291 A0A223HD43 C4MX28 B5MEK1 H9A5M2 A0A076E9A8 C1PHA8 E2S067 A0A076E5W5 E2S069 A0A1B3P5R0 A0A386NB18 A0A386H9D4 A0A0K0KG02 E2S068 E2S071 E2S072 F5CJE4 A0A345BET1 A0A212EN65 A0A140YWR4 E2S070 A0A0L7LQN7 A0A220D5Z4 A0A3S7SGT1 A0A0K1W013 A0A2H4NT96 A0A2Z4EY25 B2ZDZ6 B9VH08 A0A3G2KX47 A0A2P1JH84 S5U415 A0A0B5ITU4 A0A0L7QV02 A0A0U4DIR1 E0VHN8 A0A0H3W578 A0A0N9JVJ7 C0Z3R3 A0A0S3J2L6 A0A0C9PWC1 A0A1S5VFJ4 V9MVU4 G9J1I3 A1YWY0
PDB
6C70
E-value=1.86501e-17,
Score=211
Ontologies
GO
PANTHER
Topology
Subcellular location
Cell membrane
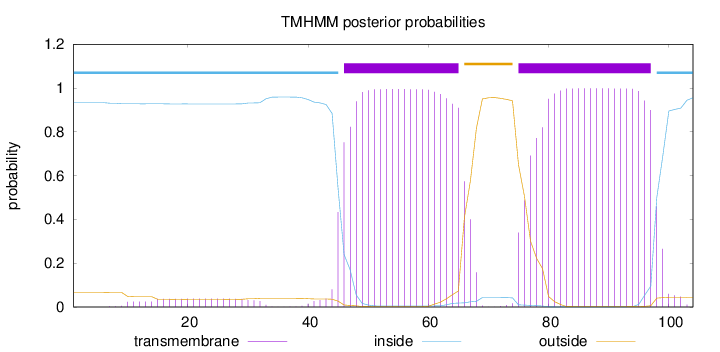
Length:
104
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
43.50017
Exp number, first 60 AAs:
15.86301
Total prob of N-in:
0.93336
POSSIBLE N-term signal
sequence
inside
1 - 45
TMhelix
46 - 65
outside
66 - 74
TMhelix
75 - 97
inside
98 - 104
Population Genetic Test Statistics
Pi
55.384704
Theta
51.920488
Tajima's D
-0.694331
CLR
422.140802
CSRT
0.195990200489976
Interpretation
Uncertain
