Gene
KWMTBOMO09566
Pre Gene Modal
BGIBMGA012832
Annotation
PREDICTED:_ras-like_protein_family_member_10B_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.453 Nuclear Reliability : 1.69
Sequence
CDS
ATGAACCGTTTCGATAAGCTTTCTATACACGATAAAGACGACGAAAATGACAGAGCAAACTGTACGAAGCCGAACAAAATATTGGAATTGGACTTGGATTACCTTGAGCGAGGCGGTGGAGGAACACTGACGACGGGCGGAGCTCCAAGCGTAGAGCCAAGCCCAGAAAGCCTTGACCTCGTCAAAGTTGTGCTACTCGGTGCTCCAGCAGTCGGAAAGACGAGTATTATCCAGCAATTTGTCTGGAGCGACTTCTCGGAAGAATACATACCTACCGATCGAAAGCACACCTTCTATCCATCGGTGATCATCAACGAGCACTTATATGAAGTTAAGATAACCGACGTTCCGGTGATCCCGTACTTTCCGATCAATTCGTACTACGAATGGGCTCACTACAGATTTTACGGACTACGGAACGCCACAGCCTACATTCTCGTGTTCGATCTGTCTAATGTCGAAACATTTCAGTACATACGAACGCTACGAGATCAAATGGTCGAGAGTCGCGACATGCGAAATGTTCCCGTGCTCGTTGTCGGCAATAAACAGGATTTGCTTTGCAGCACTACACCGTCGGCGGCGAGCGGTTTGCAACAGCTGGCAATAGCGGAAAGCGCTTGCGGCGATTCAAGGGAGAAGCGACGTAATATTGTGAATTTAGTGAGGAAACATTGGAAGTGTGGTTACGTTGAGTGCTCGGCGAAATACAATTGGAGAGTGATAGCGGTGTTCAAGGAGTTAATGGAAATGATTGATGCCCTAGAGTGGAGCGGCGGTGGTAGGAACGCGGAACCGCCTCCTGATTCATCCGCTGGTGGCGGGGCTGGTTCGCACGACACTAGATGTGTGGTTGCTTAG
Protein
MNRFDKLSIHDKDDENDRANCTKPNKILELDLDYLERGGGGTLTTGGAPSVEPSPESLDLVKVVLLGAPAVGKTSIIQQFVWSDFSEEYIPTDRKHTFYPSVIINEHLYEVKITDVPVIPYFPINSYYEWAHYRFYGLRNATAYILVFDLSNVETFQYIRTLRDQMVESRDMRNVPVLVVGNKQDLLCSTTPSAASGLQQLAIAESACGDSREKRRNIVNLVRKHWKCGYVECSAKYNWRVIAVFKELMEMIDALEWSGGGRNAEPPPDSSAGGGAGSHDTRCVVA
Summary
Uniprot
A0A437BC21
A0A2A4J5P9
A0A194Q085
A0A194R314
A0A2W1BIA3
A0A2H1V989
+ More
A0A0L7LQJ1 H9JTH0 A0A067QZY4 D2A5Q8 A0A1W4WZ59 A0A195EDF6 F4WQ26 A0A151JTY6 A0A151IHM6 A0A026W0A0 A0A2P8XLB9 E2BJ31 E2A8W4 A0A3L8E2B8 E0VF49 A0A087ZPQ4 A0A158NWL1 A0A0M9A4L1 A0A151WYI6 K7ITZ1 A0A151I5V3 A0A232EXV7 A0A310ST40 E9IW03 A0A0L8GWH5 A0A0L7QV02 A0A2P6KWS6 A0A087UK12 R7T3K1 A0A1B6MGB7 A0A3B3CYM1 A0A437CZX9 M4AU06 A0A087XRJ1 A0A3B3I2J6 A0A3P9MDV7 A0A147A9J5 A0A3Q2E586 A0A1A8UFN4 A0A1A8LRM6 A0A1A8NEU7 A0A1A8ICY3 A0A1A8DSD0 A0A1A7YG72 A0A2I4BPU8 A0A3Q3B381 A0A3Q2TFX5 A0A3B3Z551 A0A3B5MHJ5 A0A3B3ULK3 A0A3P9QD49 A0A1A8H022 A0A2U9BSV0 A0A1W4ZLS4 A0A3P8WIC0 A0A3Q2WQB3 I3K6Q6 A0A3P8R8D9 A0A3P9CMT9 A0A3P8W919 A0A3B4YNR8 A0A3Q1ELP5 A0A3P8UEW2 A0A3B4GAU1 H2U7M5
A0A0L7LQJ1 H9JTH0 A0A067QZY4 D2A5Q8 A0A1W4WZ59 A0A195EDF6 F4WQ26 A0A151JTY6 A0A151IHM6 A0A026W0A0 A0A2P8XLB9 E2BJ31 E2A8W4 A0A3L8E2B8 E0VF49 A0A087ZPQ4 A0A158NWL1 A0A0M9A4L1 A0A151WYI6 K7ITZ1 A0A151I5V3 A0A232EXV7 A0A310ST40 E9IW03 A0A0L8GWH5 A0A0L7QV02 A0A2P6KWS6 A0A087UK12 R7T3K1 A0A1B6MGB7 A0A3B3CYM1 A0A437CZX9 M4AU06 A0A087XRJ1 A0A3B3I2J6 A0A3P9MDV7 A0A147A9J5 A0A3Q2E586 A0A1A8UFN4 A0A1A8LRM6 A0A1A8NEU7 A0A1A8ICY3 A0A1A8DSD0 A0A1A7YG72 A0A2I4BPU8 A0A3Q3B381 A0A3Q2TFX5 A0A3B3Z551 A0A3B5MHJ5 A0A3B3ULK3 A0A3P9QD49 A0A1A8H022 A0A2U9BSV0 A0A1W4ZLS4 A0A3P8WIC0 A0A3Q2WQB3 I3K6Q6 A0A3P8R8D9 A0A3P9CMT9 A0A3P8W919 A0A3B4YNR8 A0A3Q1ELP5 A0A3P8UEW2 A0A3B4GAU1 H2U7M5
Pubmed
EMBL
RSAL01000093
RVE47932.1
NWSH01002962
PCG67209.1
KQ459582
KPI98992.1
+ More
KQ460878 KPJ11635.1 KZ150073 PZC74011.1 ODYU01001156 SOQ36972.1 JTDY01000319 KOB77717.1 BABH01003846 KK853054 KDR12000.1 KQ971345 EFA05040.2 KQ979074 KYN22847.1 GL888262 EGI63649.1 KQ981799 KYN35703.1 KQ977586 KYN01700.1 KK107522 EZA49340.1 PYGN01001788 PSN32810.1 GL448558 EFN84181.1 GL437663 EFN70193.1 QOIP01000001 RLU26593.1 DS235101 EEB12005.1 ADTU01002972 KQ435750 KOX76356.1 KQ982651 KYQ52827.1 KQ976434 KYM87193.1 NNAY01001711 OXU23155.1 KQ760241 OAD61295.1 GL766449 EFZ15233.1 KQ420136 KOF81174.1 KQ414731 KOC62379.1 MWRG01004149 PRD30780.1 KK120181 KFM77701.1 AMQN01015963 KB312401 ELT87273.1 GEBQ01005013 JAT34964.1 CM012445 RVE68285.1 AYCK01014547 AYCK01014548 GCES01011142 JAR75181.1 HADY01004814 HAEJ01006662 SBS47119.1 HAEF01008624 SBR46951.1 HAEH01001774 SBR67578.1 HAED01007973 HAEE01006810 SBQ94185.1 HADZ01021446 HAEA01008427 SBQ36907.1 HADW01004181 HADX01007009 SBP29241.1 HAEB01008793 HAEC01009280 SBQ77496.1 CP026251 AWP07111.1 AERX01026831
KQ460878 KPJ11635.1 KZ150073 PZC74011.1 ODYU01001156 SOQ36972.1 JTDY01000319 KOB77717.1 BABH01003846 KK853054 KDR12000.1 KQ971345 EFA05040.2 KQ979074 KYN22847.1 GL888262 EGI63649.1 KQ981799 KYN35703.1 KQ977586 KYN01700.1 KK107522 EZA49340.1 PYGN01001788 PSN32810.1 GL448558 EFN84181.1 GL437663 EFN70193.1 QOIP01000001 RLU26593.1 DS235101 EEB12005.1 ADTU01002972 KQ435750 KOX76356.1 KQ982651 KYQ52827.1 KQ976434 KYM87193.1 NNAY01001711 OXU23155.1 KQ760241 OAD61295.1 GL766449 EFZ15233.1 KQ420136 KOF81174.1 KQ414731 KOC62379.1 MWRG01004149 PRD30780.1 KK120181 KFM77701.1 AMQN01015963 KB312401 ELT87273.1 GEBQ01005013 JAT34964.1 CM012445 RVE68285.1 AYCK01014547 AYCK01014548 GCES01011142 JAR75181.1 HADY01004814 HAEJ01006662 SBS47119.1 HAEF01008624 SBR46951.1 HAEH01001774 SBR67578.1 HAED01007973 HAEE01006810 SBQ94185.1 HADZ01021446 HAEA01008427 SBQ36907.1 HADW01004181 HADX01007009 SBP29241.1 HAEB01008793 HAEC01009280 SBQ77496.1 CP026251 AWP07111.1 AERX01026831
Proteomes
UP000283053
UP000218220
UP000053268
UP000053240
UP000037510
UP000005204
+ More
UP000027135 UP000007266 UP000192223 UP000078492 UP000007755 UP000078541 UP000078542 UP000053097 UP000245037 UP000008237 UP000000311 UP000279307 UP000009046 UP000005203 UP000005205 UP000053105 UP000075809 UP000002358 UP000078540 UP000215335 UP000053454 UP000053825 UP000054359 UP000014760 UP000261560 UP000002852 UP000028760 UP000001038 UP000265180 UP000265200 UP000265020 UP000192220 UP000264800 UP000265000 UP000261480 UP000261380 UP000261500 UP000242638 UP000246464 UP000192224 UP000265120 UP000264840 UP000005207 UP000265100 UP000265160 UP000261360 UP000257200 UP000265080 UP000261460 UP000005226
UP000027135 UP000007266 UP000192223 UP000078492 UP000007755 UP000078541 UP000078542 UP000053097 UP000245037 UP000008237 UP000000311 UP000279307 UP000009046 UP000005203 UP000005205 UP000053105 UP000075809 UP000002358 UP000078540 UP000215335 UP000053454 UP000053825 UP000054359 UP000014760 UP000261560 UP000002852 UP000028760 UP000001038 UP000265180 UP000265200 UP000265020 UP000192220 UP000264800 UP000265000 UP000261480 UP000261380 UP000261500 UP000242638 UP000246464 UP000192224 UP000265120 UP000264840 UP000005207 UP000265100 UP000265160 UP000261360 UP000257200 UP000265080 UP000261460 UP000005226
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A437BC21
A0A2A4J5P9
A0A194Q085
A0A194R314
A0A2W1BIA3
A0A2H1V989
+ More
A0A0L7LQJ1 H9JTH0 A0A067QZY4 D2A5Q8 A0A1W4WZ59 A0A195EDF6 F4WQ26 A0A151JTY6 A0A151IHM6 A0A026W0A0 A0A2P8XLB9 E2BJ31 E2A8W4 A0A3L8E2B8 E0VF49 A0A087ZPQ4 A0A158NWL1 A0A0M9A4L1 A0A151WYI6 K7ITZ1 A0A151I5V3 A0A232EXV7 A0A310ST40 E9IW03 A0A0L8GWH5 A0A0L7QV02 A0A2P6KWS6 A0A087UK12 R7T3K1 A0A1B6MGB7 A0A3B3CYM1 A0A437CZX9 M4AU06 A0A087XRJ1 A0A3B3I2J6 A0A3P9MDV7 A0A147A9J5 A0A3Q2E586 A0A1A8UFN4 A0A1A8LRM6 A0A1A8NEU7 A0A1A8ICY3 A0A1A8DSD0 A0A1A7YG72 A0A2I4BPU8 A0A3Q3B381 A0A3Q2TFX5 A0A3B3Z551 A0A3B5MHJ5 A0A3B3ULK3 A0A3P9QD49 A0A1A8H022 A0A2U9BSV0 A0A1W4ZLS4 A0A3P8WIC0 A0A3Q2WQB3 I3K6Q6 A0A3P8R8D9 A0A3P9CMT9 A0A3P8W919 A0A3B4YNR8 A0A3Q1ELP5 A0A3P8UEW2 A0A3B4GAU1 H2U7M5
A0A0L7LQJ1 H9JTH0 A0A067QZY4 D2A5Q8 A0A1W4WZ59 A0A195EDF6 F4WQ26 A0A151JTY6 A0A151IHM6 A0A026W0A0 A0A2P8XLB9 E2BJ31 E2A8W4 A0A3L8E2B8 E0VF49 A0A087ZPQ4 A0A158NWL1 A0A0M9A4L1 A0A151WYI6 K7ITZ1 A0A151I5V3 A0A232EXV7 A0A310ST40 E9IW03 A0A0L8GWH5 A0A0L7QV02 A0A2P6KWS6 A0A087UK12 R7T3K1 A0A1B6MGB7 A0A3B3CYM1 A0A437CZX9 M4AU06 A0A087XRJ1 A0A3B3I2J6 A0A3P9MDV7 A0A147A9J5 A0A3Q2E586 A0A1A8UFN4 A0A1A8LRM6 A0A1A8NEU7 A0A1A8ICY3 A0A1A8DSD0 A0A1A7YG72 A0A2I4BPU8 A0A3Q3B381 A0A3Q2TFX5 A0A3B3Z551 A0A3B5MHJ5 A0A3B3ULK3 A0A3P9QD49 A0A1A8H022 A0A2U9BSV0 A0A1W4ZLS4 A0A3P8WIC0 A0A3Q2WQB3 I3K6Q6 A0A3P8R8D9 A0A3P9CMT9 A0A3P8W919 A0A3B4YNR8 A0A3Q1ELP5 A0A3P8UEW2 A0A3B4GAU1 H2U7M5
PDB
2ERX
E-value=9.71252e-15,
Score=193
Ontologies
KEGG
GO
PANTHER
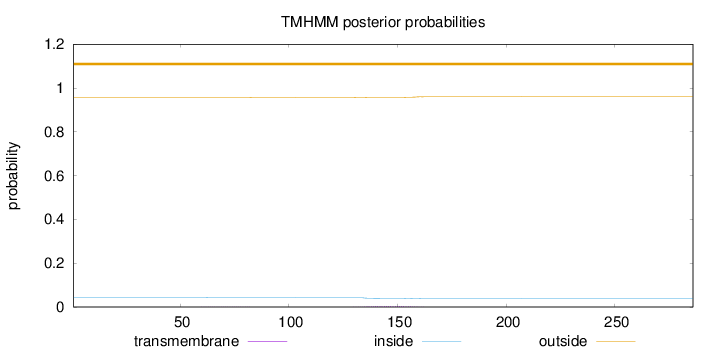
Topology
Length:
286
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.09891
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.04273
outside
1 - 286
Population Genetic Test Statistics
Pi
17.440157
Theta
21.740572
Tajima's D
-1.08154
CLR
239.951842
CSRT
0.12374381280936
Interpretation
Uncertain
