Gene
KWMTBOMO09564 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013015
Annotation
CRAL-TRIO_domain-containing_protein?_partial_[Manduca_sexta]
Location in the cell
Cytoplasmic Reliability : 3.511
Sequence
CDS
ATGACTTTGGAACAGCCAGCCGGTGAAATGTGGCAGAAGATACGCGAGGAGCTGAACGAAAATGTGGAGACCAGAGACTCCGACCTCGCCCACATCAAGGAGTGGCTGAAGAAGGAACCACATCTACCTGATGAATTCGATGACCAGCGCATCATGACCTTCCTCCGAGGTTGTAAGTTCTCCTTGGAGAAGACCAAGCGCAAGCTGGACATGTACTTCACTATGAGGACAGCTGTGCCGGAATTTTTCACGAATCGTGACGTCAAACGTCCAGAATTGCAAGAAATTTTGAGTTTAGTTCAAACTCCTGCATTACCTGGCATCACTCCCGAGGGCAGAAGGGTTGTTATTATGCGAGGACTCGATAAAGATGCTGCTACACCAAACGTTGCCGACGCTTTTAAGTTGGCCTTCATGATGGGAGACGTCAGACTTGCCGAAGAGAAGGAGGGAGTAGCAGGAGACGTGTACATTCTGGATGCTTCCGTGGCAACTCCTACGCACTTCGCCAAGTTCACACCGACCTTGGTCAAGAAATTCTTAGTTTGTGTACAGGAAGCCTATCCCGTCAGGCTGAAAGAAGTGCATGTTATCAACGTATCCCCACTTGTAGACAAAATCGTGAACTTTGTCAAGCCCTTCATAAAGGACAAGATCAAAGAAAGGATCTTCCTTCACACTGACACTAACGATCTTTACAAATATGTTCCCAAAGAAATGATGCCTGCGGAATATGGTGGTAATGCTGGTTCTATGAGCGACCTTCATGCTTCTTGGGTGAAGAAACTAGACGAGTACACAGATTGGTTTGCAGAGCAAGAGAATATCAAGGCAAACGAGGCTCTGAGACCAGGCAAGCCTACTAATTACGACGAGCTGTTCGGAATCGACGGCTCATTCCGTCAGTTGGTGATCGACTGA
Protein
MTLEQPAGEMWQKIREELNENVETRDSDLAHIKEWLKKEPHLPDEFDDQRIMTFLRGCKFSLEKTKRKLDMYFTMRTAVPEFFTNRDVKRPELQEILSLVQTPALPGITPEGRRVVIMRGLDKDAATPNVADAFKLAFMMGDVRLAEEKEGVAGDVYILDASVATPTHFAKFTPTLVKKFLVCVQEAYPVRLKEVHVINVSPLVDKIVNFVKPFIKDKIKERIFLHTDTNDLYKYVPKEMMPAEYGGNAGSMSDLHASWVKKLDEYTDWFAEQENIKANEALRPGKPTNYDELFGIDGSFRQLVID
Summary
Uniprot
A0A0S2Z3C6
H9JU03
A0A2A4JN64
A0A2W1BQL9
A0A3S2NT85
A0A2H1WHQ3
+ More
I4DIV6 I4DMP5 A0A2H4RMU2 A0A212FBP0 A0A2H4RMR9 A0A194Q1X2 A0A1E1W993 A0A2A3EKL1 A0A0N0BHJ4 A0A194R1R7 A0A087ZQ01 A0A2H4RMW4 A0A194Q6G5 A0A0L7QV98 A0A158NWN2 K7ITZ4 D2A5R3 A0A310SGC2 A0A195ECA1 A0A151WY48 A0A212FMF6 A0A3L8E2J1 A0A232ELM6 E0VHP2 B0WR71 H9JU04 J3JUN4 A0A2A4JLK9 Q178U4 A0A182G0T6 A0A0S2Z324 Q178U5 A0A1B6CGR6 F5HKV7 A0A1D6TJS6 A0A182HSP7 A0A023EQY2 A0A154PFM2 A0A182P062 A0A2H4RMR7 E2BJ28 A0A182FNW7 A0A182TL25 A0A026W000 A0A182YQJ3 A0A182UXR4 A0A182LER6 A0A182XAU9 Q7QCC6 A0A182HSP6 A0A182JE45 A0A3S2L872 A0A182JUT9 A0A182NRL3 A0A182M982 A0A182Q192 A0A182RS71 A0A182VWX1 W5JP64 A0A1B6L485 A0A2H1V2T7 A0A151I5F0 A0A151JU22 A0A336MFQ7 A0A1J1ICM2 A0A2S2R1Y2 A0A2J7Q755 A0A2H1V2R3 A0A2H8TX43 Q1HQ95 A0A3S2NHT6 A0A0S2Z350 J9JJD5 A0A194Q0B2 A0A194R296 A0A1B0CFJ4 E2A8W6 I4DM93 I4DIS4 F4WQ28 A0A1B0DII6 A0A1Y1L6E1 A0A2R7WAD2 A0A1L8DZ37 S4PY20 A0A2W1B842 A0A023F5G9 A0A212FMH3 A0A2A4JN79 A0A1B6HJI9 A0A2H4RMQ6 H9JU05 A0A194R1G6 A0A0L7LLC8
I4DIV6 I4DMP5 A0A2H4RMU2 A0A212FBP0 A0A2H4RMR9 A0A194Q1X2 A0A1E1W993 A0A2A3EKL1 A0A0N0BHJ4 A0A194R1R7 A0A087ZQ01 A0A2H4RMW4 A0A194Q6G5 A0A0L7QV98 A0A158NWN2 K7ITZ4 D2A5R3 A0A310SGC2 A0A195ECA1 A0A151WY48 A0A212FMF6 A0A3L8E2J1 A0A232ELM6 E0VHP2 B0WR71 H9JU04 J3JUN4 A0A2A4JLK9 Q178U4 A0A182G0T6 A0A0S2Z324 Q178U5 A0A1B6CGR6 F5HKV7 A0A1D6TJS6 A0A182HSP7 A0A023EQY2 A0A154PFM2 A0A182P062 A0A2H4RMR7 E2BJ28 A0A182FNW7 A0A182TL25 A0A026W000 A0A182YQJ3 A0A182UXR4 A0A182LER6 A0A182XAU9 Q7QCC6 A0A182HSP6 A0A182JE45 A0A3S2L872 A0A182JUT9 A0A182NRL3 A0A182M982 A0A182Q192 A0A182RS71 A0A182VWX1 W5JP64 A0A1B6L485 A0A2H1V2T7 A0A151I5F0 A0A151JU22 A0A336MFQ7 A0A1J1ICM2 A0A2S2R1Y2 A0A2J7Q755 A0A2H1V2R3 A0A2H8TX43 Q1HQ95 A0A3S2NHT6 A0A0S2Z350 J9JJD5 A0A194Q0B2 A0A194R296 A0A1B0CFJ4 E2A8W6 I4DM93 I4DIS4 F4WQ28 A0A1B0DII6 A0A1Y1L6E1 A0A2R7WAD2 A0A1L8DZ37 S4PY20 A0A2W1B842 A0A023F5G9 A0A212FMH3 A0A2A4JN79 A0A1B6HJI9 A0A2H4RMQ6 H9JU05 A0A194R1G6 A0A0L7LLC8
Pubmed
EMBL
KT943552
ALQ33310.1
BABH01003854
NWSH01001060
PCG72852.1
KZ150073
+ More
PZC74013.1 RSAL01000093 RVE47935.1 ODYU01008739 SOQ52601.1 AK401224 BAM17846.1 AK402563 BAM19185.1 MG434646 ATY51952.1 AGBW02009293 OWR51150.1 MG434623 ATY51929.1 KQ459582 KPI98994.1 GDQN01007486 JAT83568.1 KZ288227 PBC31792.1 KQ435750 KOX76354.1 KQ460878 KPJ11637.1 MG434662 ATY51968.1 KPI98995.1 KQ414731 KOC62381.1 ADTU01002976 KQ971345 EFA05407.1 KQ760241 OAD61294.1 KQ979074 KYN22850.1 KQ982651 KYQ52830.1 AGBW02007651 OWR54932.1 QOIP01000001 RLU26595.1 NNAY01003544 OXU19231.1 DS235171 EEB12928.1 DS232052 EDS33245.1 BABH01003855 BABH01003856 BT126949 AEE61911.1 PCG72861.1 CH477359 EAT42708.1 JXUM01037643 KQ561136 KXJ79606.1 KT943554 ALQ33312.1 EAT42707.1 GEDC01024699 JAS12599.1 AAAB01008859 EGK96859.1 APCN01000318 GAPW01002564 JAC11034.1 KQ434893 KZC10617.1 MG434634 ATY51940.1 GL448558 EFN84178.1 KK107522 EZA49342.1 EAA08040.3 RVE47936.1 AXCM01007540 AXCN02000004 ADMH02000502 ETN66182.1 GEBQ01021517 JAT18460.1 ODYU01000399 SOQ35119.1 KQ976434 KYM87194.1 KQ981799 KYN35701.1 UFQT01000848 SSX27633.1 CVRI01000047 CRK97300.1 GGMS01014782 MBY83985.1 NEVH01017447 PNF24414.1 ODYU01000400 SOQ35121.1 GFXV01006287 MBW18092.1 DQ443157 ABF51246.1 RVE47937.1 KT943548 ALQ33306.1 ABLF02025720 KPI98996.1 KPJ11639.1 AJWK01010115 GL437663 EFN70195.1 AK402411 BAM19033.1 AK401192 BAM17814.1 GL888262 EGI63651.1 AJVK01062615 GEZM01069094 JAV66647.1 KK854375 PTY15215.1 GFDF01002388 JAV11696.1 GAIX01004053 JAA88507.1 KZ150230 PZC71999.1 GBBI01002319 JAC16393.1 OWR54931.1 PCG72860.1 GECU01032846 JAS74860.1 MG434610 ATY51916.1 KPJ11638.1 JTDY01000749 KOB76021.1
PZC74013.1 RSAL01000093 RVE47935.1 ODYU01008739 SOQ52601.1 AK401224 BAM17846.1 AK402563 BAM19185.1 MG434646 ATY51952.1 AGBW02009293 OWR51150.1 MG434623 ATY51929.1 KQ459582 KPI98994.1 GDQN01007486 JAT83568.1 KZ288227 PBC31792.1 KQ435750 KOX76354.1 KQ460878 KPJ11637.1 MG434662 ATY51968.1 KPI98995.1 KQ414731 KOC62381.1 ADTU01002976 KQ971345 EFA05407.1 KQ760241 OAD61294.1 KQ979074 KYN22850.1 KQ982651 KYQ52830.1 AGBW02007651 OWR54932.1 QOIP01000001 RLU26595.1 NNAY01003544 OXU19231.1 DS235171 EEB12928.1 DS232052 EDS33245.1 BABH01003855 BABH01003856 BT126949 AEE61911.1 PCG72861.1 CH477359 EAT42708.1 JXUM01037643 KQ561136 KXJ79606.1 KT943554 ALQ33312.1 EAT42707.1 GEDC01024699 JAS12599.1 AAAB01008859 EGK96859.1 APCN01000318 GAPW01002564 JAC11034.1 KQ434893 KZC10617.1 MG434634 ATY51940.1 GL448558 EFN84178.1 KK107522 EZA49342.1 EAA08040.3 RVE47936.1 AXCM01007540 AXCN02000004 ADMH02000502 ETN66182.1 GEBQ01021517 JAT18460.1 ODYU01000399 SOQ35119.1 KQ976434 KYM87194.1 KQ981799 KYN35701.1 UFQT01000848 SSX27633.1 CVRI01000047 CRK97300.1 GGMS01014782 MBY83985.1 NEVH01017447 PNF24414.1 ODYU01000400 SOQ35121.1 GFXV01006287 MBW18092.1 DQ443157 ABF51246.1 RVE47937.1 KT943548 ALQ33306.1 ABLF02025720 KPI98996.1 KPJ11639.1 AJWK01010115 GL437663 EFN70195.1 AK402411 BAM19033.1 AK401192 BAM17814.1 GL888262 EGI63651.1 AJVK01062615 GEZM01069094 JAV66647.1 KK854375 PTY15215.1 GFDF01002388 JAV11696.1 GAIX01004053 JAA88507.1 KZ150230 PZC71999.1 GBBI01002319 JAC16393.1 OWR54931.1 PCG72860.1 GECU01032846 JAS74860.1 MG434610 ATY51916.1 KPJ11638.1 JTDY01000749 KOB76021.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053268
UP000242457
+ More
UP000053105 UP000053240 UP000005203 UP000053825 UP000005205 UP000002358 UP000007266 UP000078492 UP000075809 UP000279307 UP000215335 UP000009046 UP000002320 UP000008820 UP000069940 UP000249989 UP000007062 UP000076407 UP000075840 UP000076502 UP000075885 UP000008237 UP000069272 UP000075902 UP000053097 UP000076408 UP000075903 UP000075882 UP000075880 UP000075881 UP000075884 UP000075883 UP000075886 UP000075900 UP000075920 UP000000673 UP000078540 UP000078541 UP000183832 UP000235965 UP000007819 UP000092461 UP000000311 UP000007755 UP000092462 UP000037510
UP000053105 UP000053240 UP000005203 UP000053825 UP000005205 UP000002358 UP000007266 UP000078492 UP000075809 UP000279307 UP000215335 UP000009046 UP000002320 UP000008820 UP000069940 UP000249989 UP000007062 UP000076407 UP000075840 UP000076502 UP000075885 UP000008237 UP000069272 UP000075902 UP000053097 UP000076408 UP000075903 UP000075882 UP000075880 UP000075881 UP000075884 UP000075883 UP000075886 UP000075900 UP000075920 UP000000673 UP000078540 UP000078541 UP000183832 UP000235965 UP000007819 UP000092461 UP000000311 UP000007755 UP000092462 UP000037510
Pfam
PF00650 CRAL_TRIO
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A0S2Z3C6
H9JU03
A0A2A4JN64
A0A2W1BQL9
A0A3S2NT85
A0A2H1WHQ3
+ More
I4DIV6 I4DMP5 A0A2H4RMU2 A0A212FBP0 A0A2H4RMR9 A0A194Q1X2 A0A1E1W993 A0A2A3EKL1 A0A0N0BHJ4 A0A194R1R7 A0A087ZQ01 A0A2H4RMW4 A0A194Q6G5 A0A0L7QV98 A0A158NWN2 K7ITZ4 D2A5R3 A0A310SGC2 A0A195ECA1 A0A151WY48 A0A212FMF6 A0A3L8E2J1 A0A232ELM6 E0VHP2 B0WR71 H9JU04 J3JUN4 A0A2A4JLK9 Q178U4 A0A182G0T6 A0A0S2Z324 Q178U5 A0A1B6CGR6 F5HKV7 A0A1D6TJS6 A0A182HSP7 A0A023EQY2 A0A154PFM2 A0A182P062 A0A2H4RMR7 E2BJ28 A0A182FNW7 A0A182TL25 A0A026W000 A0A182YQJ3 A0A182UXR4 A0A182LER6 A0A182XAU9 Q7QCC6 A0A182HSP6 A0A182JE45 A0A3S2L872 A0A182JUT9 A0A182NRL3 A0A182M982 A0A182Q192 A0A182RS71 A0A182VWX1 W5JP64 A0A1B6L485 A0A2H1V2T7 A0A151I5F0 A0A151JU22 A0A336MFQ7 A0A1J1ICM2 A0A2S2R1Y2 A0A2J7Q755 A0A2H1V2R3 A0A2H8TX43 Q1HQ95 A0A3S2NHT6 A0A0S2Z350 J9JJD5 A0A194Q0B2 A0A194R296 A0A1B0CFJ4 E2A8W6 I4DM93 I4DIS4 F4WQ28 A0A1B0DII6 A0A1Y1L6E1 A0A2R7WAD2 A0A1L8DZ37 S4PY20 A0A2W1B842 A0A023F5G9 A0A212FMH3 A0A2A4JN79 A0A1B6HJI9 A0A2H4RMQ6 H9JU05 A0A194R1G6 A0A0L7LLC8
I4DIV6 I4DMP5 A0A2H4RMU2 A0A212FBP0 A0A2H4RMR9 A0A194Q1X2 A0A1E1W993 A0A2A3EKL1 A0A0N0BHJ4 A0A194R1R7 A0A087ZQ01 A0A2H4RMW4 A0A194Q6G5 A0A0L7QV98 A0A158NWN2 K7ITZ4 D2A5R3 A0A310SGC2 A0A195ECA1 A0A151WY48 A0A212FMF6 A0A3L8E2J1 A0A232ELM6 E0VHP2 B0WR71 H9JU04 J3JUN4 A0A2A4JLK9 Q178U4 A0A182G0T6 A0A0S2Z324 Q178U5 A0A1B6CGR6 F5HKV7 A0A1D6TJS6 A0A182HSP7 A0A023EQY2 A0A154PFM2 A0A182P062 A0A2H4RMR7 E2BJ28 A0A182FNW7 A0A182TL25 A0A026W000 A0A182YQJ3 A0A182UXR4 A0A182LER6 A0A182XAU9 Q7QCC6 A0A182HSP6 A0A182JE45 A0A3S2L872 A0A182JUT9 A0A182NRL3 A0A182M982 A0A182Q192 A0A182RS71 A0A182VWX1 W5JP64 A0A1B6L485 A0A2H1V2T7 A0A151I5F0 A0A151JU22 A0A336MFQ7 A0A1J1ICM2 A0A2S2R1Y2 A0A2J7Q755 A0A2H1V2R3 A0A2H8TX43 Q1HQ95 A0A3S2NHT6 A0A0S2Z350 J9JJD5 A0A194Q0B2 A0A194R296 A0A1B0CFJ4 E2A8W6 I4DM93 I4DIS4 F4WQ28 A0A1B0DII6 A0A1Y1L6E1 A0A2R7WAD2 A0A1L8DZ37 S4PY20 A0A2W1B842 A0A023F5G9 A0A212FMH3 A0A2A4JN79 A0A1B6HJI9 A0A2H4RMQ6 H9JU05 A0A194R1G6 A0A0L7LLC8
PDB
1R5L
E-value=5.19978e-19,
Score=231
Ontologies
GO
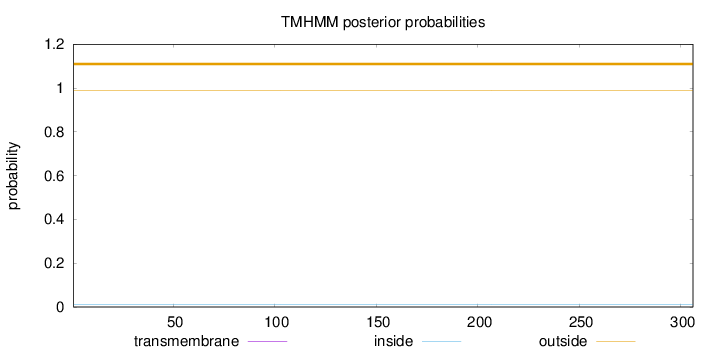
Topology
Length:
306
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000490000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00978
outside
1 - 306
Population Genetic Test Statistics
Pi
254.347485
Theta
169.158086
Tajima's D
1.42618
CLR
0.115024
CSRT
0.768161591920404
Interpretation
Uncertain
