Gene
KWMTBOMO09561 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012831
Annotation
aldo-keto_reductase_AKR2E4_[Bombyx_mori]
Full name
Aldo-keto reductase AKR2E4
Alternative Name
3-dehydroecdysone reductase
Aldo-keto reductase 2E
Aldo-keto reductase 2E
Location in the cell
Cytoplasmic Reliability : 1.798
Sequence
CDS
ATGGCGGTGCAAGCGCCTTGCATACAACTGAACGATGGTAATACTATACCCATAGTAGCTTTGGGAACTGGACGAGGAACAGCTAAGGAGTCTGACTCAATAGACGAGGTCCGCCAAGCAGTATATTGGGCAATCGAAGCTGGATACAGACATATCGACACGGCTGCCGTATACCAGGACGAGGAGCAAGTCGGTCAGGGTATAGCTGAAGCCATTGCCAATGGACTGGTCACTAGAGAAGAGCTGTTTGTTACTACTAAATTGTGGAACGACAAACACGCTCGCGACCAGGTTGTACCTGCTCTCCAAGAGTCACTCAAGAAACTTGGATTGGACTACATTGATCTATATCTCATTCATTTCCCCATAGCTACAAAGCCTGACGATTCTCCTGATAACATCGACTATCTGGAAACCTGGCAAGGCATGCAGGACGCGCGGCAGCTGGGTCTGGCCAGGTCCATTGGAGTATCCAACTTCAACGCTACGCAGATCACGAGATTGGTCAGCAACAGTTACATCAGGCCTGTTATTAACCAGATTGAGGTGAATCCTACAAACACACAAGAGCCGCTAGTGGCACACTGCCAGAGTCTCGGTATCGCCGTGATGGCGTACAGTCCGTTCGGCACTCGCTAA
Protein
MAVQAPCIQLNDGNTIPIVALGTGRGTAKESDSIDEVRQAVYWAIEAGYRHIDTAAVYQDEEQVGQGIAEAIANGLVTREELFVTTKLWNDKHARDQVVPALQESLKKLGLDYIDLYLIHFPIATKPDDSPDNIDYLETWQGMQDARQLGLARSIGVSNFNATQITRLVSNSYIRPVINQIEVNPTNTQEPLVAHCQSLGIAVMAYSPFGTR
Summary
Description
NADP-dependent oxidoreductase with high 3-dehydroecdysone reductase activity. May play a role in the regulation of molting. Has lower activity with phenylglyoxal and isatin (in vitro). Has no activity with NADH as cosubstrate. Has no activity with nitrobenzaldehyde and 3-hydroxybenzaldehyde.
Biophysicochemical Properties
0.0044 mM for 3-dehydroecdysone
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Keywords
3D-structure
Complete proteome
Lipid metabolism
NADP
Oxidoreductase
Reference proteome
Steroid metabolism
Feature
chain Aldo-keto reductase AKR2E4
Uniprot
H9JTG9
A0A291P0T7
A0A2A4JMB2
A0A2W1BC51
A0A212FML6
A0A2H1VAA3
+ More
A0A2H1WAV6 B2DBM2 A0A291P0X3 A0A3S2LZY0 A0A194R6Z8 A0A194Q090 G9F9G0 A0A0L7LKQ3 A0A2H1VGR3 A0A2H1VAB6 A0A2A4JM15 I4DP16 A0A291P0W3 A0A291P0U1 A0A437BCA7 A0A2W1BGS7 Q8WRT0 U5KC22 A0A1L8D6L9 U5KBY4 A0A3S2TM02 A0A291P0Y9 A0A291P0U2 A0A0F6Q2W6 A0A2A4IYA3 D2SNM4 A0A2H1V815 G9F9F4 A0A3S2LEX1 A0A2W1BLQ6 A0A194PSF7 S4PY90 A0A291P0Y7 A0A291P0V1 S4P5J6 G9D4S7 A0A194PKB3 A0A194QT42 A0A291P0Z6 G9D4S4 H9IVT6 A0A194QCQ5 G9D4S6 G9D4T5 A0A1L8D6N6 A0A2A4JDC7 A0A3S2NPB4 G9D4T3 A0A3S2TJM5 G9D4T6 G9D4T2 A0A194RJ39 A0A1I1AK31 G9D4S9 G9D4T1 D9HQ59 D2A296 G9F9F8 G9D4T4 A0A2H1WKT0 G9D4S5 A0A3E0E726 A0A212F4I6 A0A212F244 L7M8R1 A0A3S2M2X1 G9D4T0 A0A2A4IZB1 A0A1L8D6N3 A0A232EMX7 A0A131YR32 A0A1I7DMB9 A0A224YH56 A0A2W7RB29 A0A0N1IF51 A0A2A4K2R0 A0A2A4IW06 A0A212FE89 A0A437BH13 A0A088AGI5 A0A131XMC9 A0A194PUL9 A0A1B2G267 I4DKQ3 A0A2A4JAY8 G3MFC0 D6WCX8 V9IFI2 U5KCB8 A0A212FE35 A0A327PS09 A0A212EU63
A0A2H1WAV6 B2DBM2 A0A291P0X3 A0A3S2LZY0 A0A194R6Z8 A0A194Q090 G9F9G0 A0A0L7LKQ3 A0A2H1VGR3 A0A2H1VAB6 A0A2A4JM15 I4DP16 A0A291P0W3 A0A291P0U1 A0A437BCA7 A0A2W1BGS7 Q8WRT0 U5KC22 A0A1L8D6L9 U5KBY4 A0A3S2TM02 A0A291P0Y9 A0A291P0U2 A0A0F6Q2W6 A0A2A4IYA3 D2SNM4 A0A2H1V815 G9F9F4 A0A3S2LEX1 A0A2W1BLQ6 A0A194PSF7 S4PY90 A0A291P0Y7 A0A291P0V1 S4P5J6 G9D4S7 A0A194PKB3 A0A194QT42 A0A291P0Z6 G9D4S4 H9IVT6 A0A194QCQ5 G9D4S6 G9D4T5 A0A1L8D6N6 A0A2A4JDC7 A0A3S2NPB4 G9D4T3 A0A3S2TJM5 G9D4T6 G9D4T2 A0A194RJ39 A0A1I1AK31 G9D4S9 G9D4T1 D9HQ59 D2A296 G9F9F8 G9D4T4 A0A2H1WKT0 G9D4S5 A0A3E0E726 A0A212F4I6 A0A212F244 L7M8R1 A0A3S2M2X1 G9D4T0 A0A2A4IZB1 A0A1L8D6N3 A0A232EMX7 A0A131YR32 A0A1I7DMB9 A0A224YH56 A0A2W7RB29 A0A0N1IF51 A0A2A4K2R0 A0A2A4IW06 A0A212FE89 A0A437BH13 A0A088AGI5 A0A131XMC9 A0A194PUL9 A0A1B2G267 I4DKQ3 A0A2A4JAY8 G3MFC0 D6WCX8 V9IFI2 U5KCB8 A0A212FE35 A0A327PS09 A0A212EU63
EC Number
1.1.1.-
Pubmed
EMBL
AB701383
MF687576
ATJ44502.1
NWSH01001060
PCG72858.1
KZ150230
+ More
PZC72001.1 AGBW02007651 OWR54929.1 ODYU01001496 SOQ37780.1 ODYU01007438 SOQ50215.1 AB264706 BAG30781.1 MF687617 ATJ44543.1 RSAL01000093 RVE47941.1 KQ460878 KPJ11641.1 KQ459582 KPI98997.1 JN033705 AEW46858.1 JTDY01000749 KOB76022.1 ODYU01002492 SOQ40039.1 SOQ37779.1 PCG72859.1 AK403246 BAM19656.1 MF687607 ATJ44533.1 MF687570 ATJ44496.1 RVE47938.1 PZC72000.1 AF409102 AAL73387.1 KC007348 AGQ45613.1 GEYN01000020 JAV02109.1 KC007350 AGQ45615.1 RSAL01000058 RVE49735.1 MF687608 ATJ44534.1 MF687572 ATJ44498.1 KP237872 AKD01725.1 NWSH01005088 PCG64446.1 EZ407153 ACX53715.1 ODYU01001161 SOQ36985.1 JN033699 JN033700 RSAL01000051 AEW46852.1 AEW46853.1 RVE50216.1 RSAL01000168 RVE45243.1 KZ150094 PZC73640.1 KQ459593 KPI96371.1 GAIX01003703 JAA88857.1 MF687609 ATJ44535.1 MF687580 ATJ44506.1 GAIX01005504 JAA87056.1 JF777330 AEU11675.1 KQ459602 KPI93169.1 KQ461155 KPJ08170.1 MF687618 ATJ44544.1 JF777327 AEU11672.1 BABH01038068 BABH01038069 BABH01038070 KQ459232 KPJ02765.1 JF777329 AEU11674.1 JF777338 AEU11683.1 GEYN01000021 JAV02108.1 NWSH01001964 PCG69554.1 RSAL01000916 RVE41050.1 JF777336 AEU11681.1 RVE47940.1 JF777339 AEU11684.1 JF777335 AEU11680.1 KQ460152 KPJ17355.1 FOKK01000008 SFB36800.1 JF777332 AEU11677.1 JF777334 AEU11679.1 HM023814 ADJ58547.1 KQ971338 EFA02158.1 JN033703 AEW46856.1 JF777337 AEU11682.1 ODYU01009309 SOQ53638.1 JF777328 AEU11673.1 QUNF01000002 REG92796.1 AGBW02010353 OWR48651.1 AGBW02010783 OWR47810.1 GACK01005535 JAA59499.1 RVE49736.1 JF777333 AEU11678.1 NWSH01004698 PCG64758.1 GEYN01000016 JAV02113.1 NNAY01003257 OXU19723.1 GEDV01008001 JAP80556.1 FPBF01000007 SFU12766.1 GFPF01005872 MAA17018.1 QKZU01000009 PZX55450.1 KQ460773 KPJ12435.1 NWSH01000243 PCG78063.1 NWSH01006101 PCG63656.1 AGBW02008974 OWR52018.1 RVE49724.1 GEFH01001890 JAP66691.1 KPI96444.1 KU233524 ANZ02995.1 AK401871 BAM18493.1 NWSH01002278 PCG68704.1 JO840571 AEO32188.1 KQ971311 EEZ99072.1 JR040675 AEY59236.1 KC007356 AGQ45621.1 OWR52019.1 QLLK01000002 RAI94101.1 AGBW02012481 OWR44997.1
PZC72001.1 AGBW02007651 OWR54929.1 ODYU01001496 SOQ37780.1 ODYU01007438 SOQ50215.1 AB264706 BAG30781.1 MF687617 ATJ44543.1 RSAL01000093 RVE47941.1 KQ460878 KPJ11641.1 KQ459582 KPI98997.1 JN033705 AEW46858.1 JTDY01000749 KOB76022.1 ODYU01002492 SOQ40039.1 SOQ37779.1 PCG72859.1 AK403246 BAM19656.1 MF687607 ATJ44533.1 MF687570 ATJ44496.1 RVE47938.1 PZC72000.1 AF409102 AAL73387.1 KC007348 AGQ45613.1 GEYN01000020 JAV02109.1 KC007350 AGQ45615.1 RSAL01000058 RVE49735.1 MF687608 ATJ44534.1 MF687572 ATJ44498.1 KP237872 AKD01725.1 NWSH01005088 PCG64446.1 EZ407153 ACX53715.1 ODYU01001161 SOQ36985.1 JN033699 JN033700 RSAL01000051 AEW46852.1 AEW46853.1 RVE50216.1 RSAL01000168 RVE45243.1 KZ150094 PZC73640.1 KQ459593 KPI96371.1 GAIX01003703 JAA88857.1 MF687609 ATJ44535.1 MF687580 ATJ44506.1 GAIX01005504 JAA87056.1 JF777330 AEU11675.1 KQ459602 KPI93169.1 KQ461155 KPJ08170.1 MF687618 ATJ44544.1 JF777327 AEU11672.1 BABH01038068 BABH01038069 BABH01038070 KQ459232 KPJ02765.1 JF777329 AEU11674.1 JF777338 AEU11683.1 GEYN01000021 JAV02108.1 NWSH01001964 PCG69554.1 RSAL01000916 RVE41050.1 JF777336 AEU11681.1 RVE47940.1 JF777339 AEU11684.1 JF777335 AEU11680.1 KQ460152 KPJ17355.1 FOKK01000008 SFB36800.1 JF777332 AEU11677.1 JF777334 AEU11679.1 HM023814 ADJ58547.1 KQ971338 EFA02158.1 JN033703 AEW46856.1 JF777337 AEU11682.1 ODYU01009309 SOQ53638.1 JF777328 AEU11673.1 QUNF01000002 REG92796.1 AGBW02010353 OWR48651.1 AGBW02010783 OWR47810.1 GACK01005535 JAA59499.1 RVE49736.1 JF777333 AEU11678.1 NWSH01004698 PCG64758.1 GEYN01000016 JAV02113.1 NNAY01003257 OXU19723.1 GEDV01008001 JAP80556.1 FPBF01000007 SFU12766.1 GFPF01005872 MAA17018.1 QKZU01000009 PZX55450.1 KQ460773 KPJ12435.1 NWSH01000243 PCG78063.1 NWSH01006101 PCG63656.1 AGBW02008974 OWR52018.1 RVE49724.1 GEFH01001890 JAP66691.1 KPI96444.1 KU233524 ANZ02995.1 AK401871 BAM18493.1 NWSH01002278 PCG68704.1 JO840571 AEO32188.1 KQ971311 EEZ99072.1 JR040675 AEY59236.1 KC007356 AGQ45621.1 OWR52019.1 QLLK01000002 RAI94101.1 AGBW02012481 OWR44997.1
Proteomes
Pfam
PF00248 Aldo_ket_red
Interpro
SUPFAM
SSF51430
SSF51430
Gene 3D
ProteinModelPortal
H9JTG9
A0A291P0T7
A0A2A4JMB2
A0A2W1BC51
A0A212FML6
A0A2H1VAA3
+ More
A0A2H1WAV6 B2DBM2 A0A291P0X3 A0A3S2LZY0 A0A194R6Z8 A0A194Q090 G9F9G0 A0A0L7LKQ3 A0A2H1VGR3 A0A2H1VAB6 A0A2A4JM15 I4DP16 A0A291P0W3 A0A291P0U1 A0A437BCA7 A0A2W1BGS7 Q8WRT0 U5KC22 A0A1L8D6L9 U5KBY4 A0A3S2TM02 A0A291P0Y9 A0A291P0U2 A0A0F6Q2W6 A0A2A4IYA3 D2SNM4 A0A2H1V815 G9F9F4 A0A3S2LEX1 A0A2W1BLQ6 A0A194PSF7 S4PY90 A0A291P0Y7 A0A291P0V1 S4P5J6 G9D4S7 A0A194PKB3 A0A194QT42 A0A291P0Z6 G9D4S4 H9IVT6 A0A194QCQ5 G9D4S6 G9D4T5 A0A1L8D6N6 A0A2A4JDC7 A0A3S2NPB4 G9D4T3 A0A3S2TJM5 G9D4T6 G9D4T2 A0A194RJ39 A0A1I1AK31 G9D4S9 G9D4T1 D9HQ59 D2A296 G9F9F8 G9D4T4 A0A2H1WKT0 G9D4S5 A0A3E0E726 A0A212F4I6 A0A212F244 L7M8R1 A0A3S2M2X1 G9D4T0 A0A2A4IZB1 A0A1L8D6N3 A0A232EMX7 A0A131YR32 A0A1I7DMB9 A0A224YH56 A0A2W7RB29 A0A0N1IF51 A0A2A4K2R0 A0A2A4IW06 A0A212FE89 A0A437BH13 A0A088AGI5 A0A131XMC9 A0A194PUL9 A0A1B2G267 I4DKQ3 A0A2A4JAY8 G3MFC0 D6WCX8 V9IFI2 U5KCB8 A0A212FE35 A0A327PS09 A0A212EU63
A0A2H1WAV6 B2DBM2 A0A291P0X3 A0A3S2LZY0 A0A194R6Z8 A0A194Q090 G9F9G0 A0A0L7LKQ3 A0A2H1VGR3 A0A2H1VAB6 A0A2A4JM15 I4DP16 A0A291P0W3 A0A291P0U1 A0A437BCA7 A0A2W1BGS7 Q8WRT0 U5KC22 A0A1L8D6L9 U5KBY4 A0A3S2TM02 A0A291P0Y9 A0A291P0U2 A0A0F6Q2W6 A0A2A4IYA3 D2SNM4 A0A2H1V815 G9F9F4 A0A3S2LEX1 A0A2W1BLQ6 A0A194PSF7 S4PY90 A0A291P0Y7 A0A291P0V1 S4P5J6 G9D4S7 A0A194PKB3 A0A194QT42 A0A291P0Z6 G9D4S4 H9IVT6 A0A194QCQ5 G9D4S6 G9D4T5 A0A1L8D6N6 A0A2A4JDC7 A0A3S2NPB4 G9D4T3 A0A3S2TJM5 G9D4T6 G9D4T2 A0A194RJ39 A0A1I1AK31 G9D4S9 G9D4T1 D9HQ59 D2A296 G9F9F8 G9D4T4 A0A2H1WKT0 G9D4S5 A0A3E0E726 A0A212F4I6 A0A212F244 L7M8R1 A0A3S2M2X1 G9D4T0 A0A2A4IZB1 A0A1L8D6N3 A0A232EMX7 A0A131YR32 A0A1I7DMB9 A0A224YH56 A0A2W7RB29 A0A0N1IF51 A0A2A4K2R0 A0A2A4IW06 A0A212FE89 A0A437BH13 A0A088AGI5 A0A131XMC9 A0A194PUL9 A0A1B2G267 I4DKQ3 A0A2A4JAY8 G3MFC0 D6WCX8 V9IFI2 U5KCB8 A0A212FE35 A0A327PS09 A0A212EU63
PDB
3WCZ
E-value=6.57817e-119,
Score=1090
Ontologies
PATHWAY
00040
Pentose and glucuronate interconversions - Bombyx mori (domestic silkworm)
00051 Fructose and mannose metabolism - Bombyx mori (domestic silkworm)
00052 Galactose metabolism - Bombyx mori (domestic silkworm)
00561 Glycerolipid metabolism - Bombyx mori (domestic silkworm)
00790 Folate biosynthesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00051 Fructose and mannose metabolism - Bombyx mori (domestic silkworm)
00052 Galactose metabolism - Bombyx mori (domestic silkworm)
00561 Glycerolipid metabolism - Bombyx mori (domestic silkworm)
00790 Folate biosynthesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
PANTHER
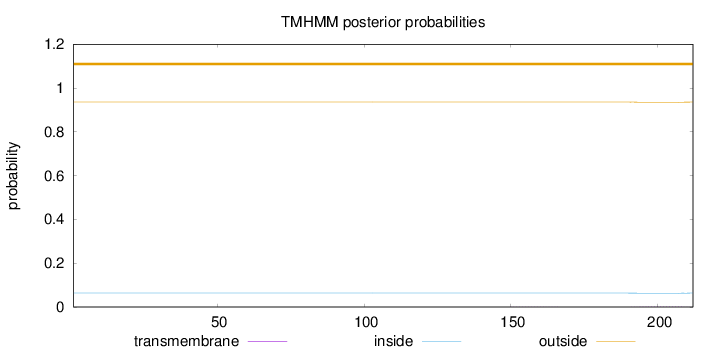
Topology
Length:
212
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0283
Exp number, first 60 AAs:
0.0008
Total prob of N-in:
0.06375
outside
1 - 212
Population Genetic Test Statistics
Pi
185.476456
Theta
18.012847
Tajima's D
0.149759
CLR
1.781719
CSRT
0.413779311034448
Interpretation
Uncertain
