Gene
KWMTBOMO09558
Pre Gene Modal
BGIBMGA012830
Annotation
PREDICTED:_phosphatidylinositol_4-kinase_alpha_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.115 Nuclear Reliability : 1.03 PlasmaMembrane Reliability : 1.258
Sequence
CDS
ATGTACAATCATTGGCATCCAAGCCAATCACTGTACCACTATACAAGAAAAAATACAGTGATTGGCACCAAAGAAATTACCGGTTCAAAACTGGAGCCCCGACCTCCGTTCGTGGCCCCGCACGCGGTGTGGGTCCGCTACTTGTGCGAAGTGGCGGAGACCGCCAAATACAACAGCCAGGAGAAGGTCGAGATGCTGGCGAGCCTGCTGCACCGATCTTTGCCGATCACTGTGGGCGATTACCGCGATCACATAAACAGACACGTCGAAGCAGTCGGAGTCAGATTCAAACTCCTCTCGTGCGGTCTCTCTTTACTGCAAGGCGACATACTGCCGCGCTCGCTGGCCAGGAACATACTCCGCGAACGCGTGTACAGCGCCTGCTTGGACTACTTCTGCCGAGGGATCCAGTGCCCGTCGAAAAGCTCCGGAGCTTTACGAGAAGATATCATATCGTTGATAAAGTTCTGGCAGCTCATGCACTCGGATAAGAAGTACTTGAAGAGCAGCGATATAGGTGGTGAATACGAGCTGATGGGTCCGAGCAGCCAGCAGAGCCTCTACACGTCGAACTCGCCCACGGACAACAGGTCGTCGGTGAACAGCAGCGACATCGGCAGCCGTCAGACCACGGGCTGGATCAACACGGTACCAATGTCCACGACCACGCTGAGCAGCGGAGCGGGTAGCATAGCGGGGAAACGATCTAACCGAAGCGTCAAGCCACCCGTCAAGCAACAGGACACGTACGTCAAGGACTACGTCAGGAAAAGGAATCTTATATTGGAGCTTATGGCGGTCGAGATAGAGTTCTTGGTGACGTGGCACAACCCTCACGGGCGGCCCGAGCAGGCCGTCCCGGGTGAGGACAACATCGCCACCTGGCGCTCCAAGCCCAACACCGAGCGGACCTGGAGGGACCACGCCAAGCTGGCCTGGGAAATCAGTCCCGTTCTCGCCGTCTTCATGCCGGAGAGGCTAAAGAACGACGGTATAGTGGCGGAGGTCAGACGCAAAGTCCAAGCGCAGCCGACCGCGGTGTGCCACGTCCCCCAGGCCCTGAAGTATCTGGTCACCACCGAAACCTTACTCAACGATGCGCAAGAGCTGGTACACATGATAACCTGGGCCCGGGTCAGTCCGGTGGAGGCCCTGTCGTACTTCAGCCGGCAGTACCCGCCGCACCCGCTGTCGGCGCAGGCGGCCGTCAACACGCTGACGTCATACCCCTCCTCGACCCTCCTCCTGTACATTCCGCAGCTTGTGCAGGCGCTGCGGCACGACACGATGGGATACGTGGCGGAATTGATAAAATCACTGGCGAAGAAATCACAAGTCGTAGCGCACCAGCTGATATGGAACATGCACACGAACATGTACACAGACGAGGAGATGCATAATAAAGATCCGGCCCTATTCGACATCCTCGAGTCGTTGACGAAGAGCATAGTGGATTCCCTGTCGGGACCCGCGAAGGCGTTCTACGAAAGGGAATTCGATTTCTTCGGTCAGATCACGAACATCAGCGGAATCATCAGACCGTACCCCAAGGGAGCGGAGAGGAAGCGAGCCTGCTTGGAAGCGCTGAAGAACGTTAAGGTGCAGCCGGGGTGCTATCTACCCTCCAATCCGGACTCTATGGTCATCGATATAGACTATAAAAGCGGTACTCCCATGCAGAGCGCCGCGAAGGCTCCGTACTTAGCGCGGTTCAGAGTCCGAAAGTACGGGATCGCGGAGATGGAGTCCCTCGCCATGGCCATCGCGTCGGGCGAGGAGCCGCCTGCGGAGGAAGGCAAGGACCGGTACACGTCGATAGCGGCGGAGACCTGGCAGGCGGCCATATTTAAAGTGGGCGATGACGTCAGACAAGACATGCTGGCCCTGCAAGTGATATCGCTGTTCAAGAACATATTCCAGCAAGTCGGCCTGGACTTGTACCTGTTTCCGTACAAAGTCGTCGCCACCGCGCCCGGGAGCGGCGTTATAGAATGCGTGCCCAACGCCAAGTCGCGTGATCAGCTGGGCAGGCAGACGGACTTCGGACTCTACGAATATTTTATAAAGAAATACGGCGACGAGACCACGAGAGAGTTTCAGGCGGCGAGAAGGTGCTTCGTGACCTCGATGGCCGCGTACAGCGTCATCGGCTTCCTGCTGCAGATCAAGGATCGGCACAACGGGAACATCATGCTGGACACGGACGGACACATCATACACATAGATTTCGGATTCATGTTCGAGTCATCGCCCGGCGGGAACCTTGGCTTCGAGCCCGACATCAAGCTGACCGACGAGATGGTGATGGTGATGGGCGGCAAGATGGAGGCGCCGCCGTTCCGCTGGTTCTGTGAACTGTGCGTGCAGGCCTTCTTGGCTGTCAGACCATACCAAGAAGCTATAATATCTATGGTGTCGCTGATGCTGGACACCGGTCTCCCGTGCTTCCGAGGGCAGACGATACGTCACCTTCGCTCGCGTTTCGCGCCCAACTCTACCGAGAAGGAGGCGGCGGCGTACATGCTCTCTATCATACGTAACTCGTTCTTGAACTTTAGAACAAGGACGTACGATATAATACAGTATTACCAGAACCAGATACCGTATTGA
Protein
MYNHWHPSQSLYHYTRKNTVIGTKEITGSKLEPRPPFVAPHAVWVRYLCEVAETAKYNSQEKVEMLASLLHRSLPITVGDYRDHINRHVEAVGVRFKLLSCGLSLLQGDILPRSLARNILRERVYSACLDYFCRGIQCPSKSSGALREDIISLIKFWQLMHSDKKYLKSSDIGGEYELMGPSSQQSLYTSNSPTDNRSSVNSSDIGSRQTTGWINTVPMSTTTLSSGAGSIAGKRSNRSVKPPVKQQDTYVKDYVRKRNLILELMAVEIEFLVTWHNPHGRPEQAVPGEDNIATWRSKPNTERTWRDHAKLAWEISPVLAVFMPERLKNDGIVAEVRRKVQAQPTAVCHVPQALKYLVTTETLLNDAQELVHMITWARVSPVEALSYFSRQYPPHPLSAQAAVNTLTSYPSSTLLLYIPQLVQALRHDTMGYVAELIKSLAKKSQVVAHQLIWNMHTNMYTDEEMHNKDPALFDILESLTKSIVDSLSGPAKAFYEREFDFFGQITNISGIIRPYPKGAERKRACLEALKNVKVQPGCYLPSNPDSMVIDIDYKSGTPMQSAAKAPYLARFRVRKYGIAEMESLAMAIASGEEPPAEEGKDRYTSIAAETWQAAIFKVGDDVRQDMLALQVISLFKNIFQQVGLDLYLFPYKVVATAPGSGVIECVPNAKSRDQLGRQTDFGLYEYFIKKYGDETTREFQAARRCFVTSMAAYSVIGFLLQIKDRHNGNIMLDTDGHIIHIDFGFMFESSPGGNLGFEPDIKLTDEMVMVMGGKMEAPPFRWFCELCVQAFLAVRPYQEAIISMVSLMLDTGLPCFRGQTIRHLRSRFAPNSTEKEAAAYMLSIIRNSFLNFRTRTYDIIQYYQNQIPY
Summary
Similarity
Belongs to the PI3/PI4-kinase family.
Belongs to the peptidase C2 family.
Belongs to the peptidase C2 family.
Uniprot
A0A212FME8
H9JTG8
A0A3S2TCX0
A0A067QIG5
E0W3P8
A0A069DZT9
+ More
A0A023F555 A0A1B6HW70 A0A1B6LF78 A0A0T6BCD2 D2A3W3 A0A2A4JVW0 A0A0A9ZGW4 A0A0A9ZFG5 A0A2H8TZ06 A0A2S2Q5K8 J9JMX5 A0A2J7QZA3 A0A1Y1M0R4 A0A0C9QK07 E2AJ31 A0A0J7L641 U4U5D1 A0A088A932 A0A2A3EB47 A0A195AUW7 E2B3H5 A0A158NNB0 A0A195FY16 A0A195CS11 A0A151IT87 A0A026WVJ6 A0A3L8DIY4 A0A1Q3G3S1 A0A182NV87 B0W0P5 A0A182QGM5 A0A182P1W5 A0A182TSY5 Q7Q237 A0A182RUL6 A0A182I5Q9 W4VRQ0 A0A182JI18 A0A182W9I8 A0A182JW38 A0A1S4EV63 Q17PY3 A0A182GH84 A0A182YIY1 A0A084W1F9 A0A1S4EV62 A0A182MWN4 A0A2M4A7Q3 A0A2M4B8D4 A0A2M4B873 T1J2J9 A0A2M4A6S0 A0A182FVH4 A0A2M4B8D6 W5JJV1 B7QKB8 A0A131XVJ2 V5IIQ8 A0A293MAF0 A0A182X0H0 A0A1I8Q522 A0A1I8Q510 A0A2P6KWY6 V5GI35 A0A0A1X897 A0A0A1WMM2 A0A293LPG1 A0A1A9ULE3 A0A232ER76 A0A1A9XE14 A0A1B0BVY8 A0A1A9W400 A0A1B0AC15 W8BMJ7 A0A1B0FEG5 L7LVG2 A0A131YYL1 K7J6G1 A0A0K8WFR5 A0A0K8TYY2 A0A1E1XB13 A0A0L0BYP1 W8C6T2 A0A034V5U8 A0A1I8M8W2 A0A131XIY6 A0A0M4ETM9 A0A224Z5V5 B4NCK6 B4L8F2 A0A1E1XQJ6 A0A0Q9XGA2 A0A0R3P3H5 A0A1J1I3I5
A0A023F555 A0A1B6HW70 A0A1B6LF78 A0A0T6BCD2 D2A3W3 A0A2A4JVW0 A0A0A9ZGW4 A0A0A9ZFG5 A0A2H8TZ06 A0A2S2Q5K8 J9JMX5 A0A2J7QZA3 A0A1Y1M0R4 A0A0C9QK07 E2AJ31 A0A0J7L641 U4U5D1 A0A088A932 A0A2A3EB47 A0A195AUW7 E2B3H5 A0A158NNB0 A0A195FY16 A0A195CS11 A0A151IT87 A0A026WVJ6 A0A3L8DIY4 A0A1Q3G3S1 A0A182NV87 B0W0P5 A0A182QGM5 A0A182P1W5 A0A182TSY5 Q7Q237 A0A182RUL6 A0A182I5Q9 W4VRQ0 A0A182JI18 A0A182W9I8 A0A182JW38 A0A1S4EV63 Q17PY3 A0A182GH84 A0A182YIY1 A0A084W1F9 A0A1S4EV62 A0A182MWN4 A0A2M4A7Q3 A0A2M4B8D4 A0A2M4B873 T1J2J9 A0A2M4A6S0 A0A182FVH4 A0A2M4B8D6 W5JJV1 B7QKB8 A0A131XVJ2 V5IIQ8 A0A293MAF0 A0A182X0H0 A0A1I8Q522 A0A1I8Q510 A0A2P6KWY6 V5GI35 A0A0A1X897 A0A0A1WMM2 A0A293LPG1 A0A1A9ULE3 A0A232ER76 A0A1A9XE14 A0A1B0BVY8 A0A1A9W400 A0A1B0AC15 W8BMJ7 A0A1B0FEG5 L7LVG2 A0A131YYL1 K7J6G1 A0A0K8WFR5 A0A0K8TYY2 A0A1E1XB13 A0A0L0BYP1 W8C6T2 A0A034V5U8 A0A1I8M8W2 A0A131XIY6 A0A0M4ETM9 A0A224Z5V5 B4NCK6 B4L8F2 A0A1E1XQJ6 A0A0Q9XGA2 A0A0R3P3H5 A0A1J1I3I5
Pubmed
22118469
19121390
24845553
20566863
26334808
25474469
+ More
18362917 19820115 25401762 28004739 20798317 23537049 21347285 24508170 30249741 12364791 14747013 17210077 17510324 26483478 25244985 24438588 20920257 23761445 25765539 25830018 28648823 24495485 25576852 26830274 20075255 28503490 26108605 25348373 25315136 28049606 28797301 17994087 29209593 15632085
18362917 19820115 25401762 28004739 20798317 23537049 21347285 24508170 30249741 12364791 14747013 17210077 17510324 26483478 25244985 24438588 20920257 23761445 25765539 25830018 28648823 24495485 25576852 26830274 20075255 28503490 26108605 25348373 25315136 28049606 28797301 17994087 29209593 15632085
EMBL
AGBW02007651
OWR54925.1
BABH01003858
BABH01003859
BABH01003860
RSAL01000342
+ More
RVE42389.1 KK853329 KDR08422.1 DS235882 EEB20254.1 GBGD01000060 JAC88829.1 GBBI01002518 JAC16194.1 GECU01028793 GECU01025296 GECU01016951 JAS78913.1 JAS82410.1 JAS90755.1 GEBQ01017648 JAT22329.1 LJIG01001952 KRT84988.1 KQ971348 EFA04876.1 NWSH01000564 PCG75623.1 GBHO01002289 GBHO01000336 JAG41315.1 JAG43268.1 GBHO01000337 GBHO01000335 JAG43267.1 JAG43269.1 GFXV01007324 MBW19129.1 GGMS01003842 MBY73045.1 ABLF02021159 ABLF02066729 NEVH01009070 PNF33920.1 GEZM01043027 JAV79373.1 GBYB01003874 JAG73641.1 GL439932 EFN66563.1 LBMM01000586 KMQ98023.1 KB632145 ERL89099.1 KZ288318 PBC28281.1 KQ976738 KYM75852.1 GL445346 EFN89727.1 ADTU01021199 ADTU01021200 KQ981208 KYN44729.1 KQ977372 KYN03282.1 KQ981037 KYN10162.1 KK107109 EZA59099.1 QOIP01000008 RLU19889.1 GFDL01000604 JAV34441.1 DS231818 EDS41347.1 AXCN02001805 AAAB01008979 EAA13769.4 APCN01003485 GANO01000560 JAB59311.1 CH477188 EAT48811.1 JXUM01001263 JXUM01001264 KQ560119 KXJ84380.1 ATLV01019337 KE525268 KFB44053.1 AXCM01006588 GGFK01003502 MBW36823.1 GGFJ01000109 MBW49250.1 GGFJ01000108 MBW49249.1 JH431806 GGFK01003134 MBW36455.1 GGFJ01000110 MBW49251.1 ADMH02001311 ETN63054.1 ABJB010908985 DS958725 EEC19290.1 GEFM01006110 JAP69686.1 GANP01002762 JAB81706.1 GFWV01013093 MAA37822.1 MWRG01004097 PRD30826.1 GANP01014563 JAB69905.1 GBXI01007327 JAD06965.1 GBXI01014386 JAC99905.1 GFWV01005852 MAA30582.1 NNAY01002664 OXU20832.1 JXJN01021533 JXJN01021534 GAMC01008352 GAMC01008349 JAB98203.1 CCAG010016028 GACK01009462 JAA55572.1 GEDV01004284 JAP84273.1 GDHF01002387 JAI49927.1 GDHF01032642 JAI19672.1 GFAC01002912 JAT96276.1 JRES01001143 KNC25163.1 GAMC01008354 GAMC01008351 JAB98204.1 GAKP01021073 JAC37879.1 GEFH01002084 JAP66497.1 CP012528 ALC49915.1 GFPF01010546 MAA21692.1 CH964239 EDW82565.1 CH933815 EDW07927.1 GFAA01002135 JAU01300.1 KRG07526.1 CH379063 KRT06173.1 CVRI01000040 CRK94872.1
RVE42389.1 KK853329 KDR08422.1 DS235882 EEB20254.1 GBGD01000060 JAC88829.1 GBBI01002518 JAC16194.1 GECU01028793 GECU01025296 GECU01016951 JAS78913.1 JAS82410.1 JAS90755.1 GEBQ01017648 JAT22329.1 LJIG01001952 KRT84988.1 KQ971348 EFA04876.1 NWSH01000564 PCG75623.1 GBHO01002289 GBHO01000336 JAG41315.1 JAG43268.1 GBHO01000337 GBHO01000335 JAG43267.1 JAG43269.1 GFXV01007324 MBW19129.1 GGMS01003842 MBY73045.1 ABLF02021159 ABLF02066729 NEVH01009070 PNF33920.1 GEZM01043027 JAV79373.1 GBYB01003874 JAG73641.1 GL439932 EFN66563.1 LBMM01000586 KMQ98023.1 KB632145 ERL89099.1 KZ288318 PBC28281.1 KQ976738 KYM75852.1 GL445346 EFN89727.1 ADTU01021199 ADTU01021200 KQ981208 KYN44729.1 KQ977372 KYN03282.1 KQ981037 KYN10162.1 KK107109 EZA59099.1 QOIP01000008 RLU19889.1 GFDL01000604 JAV34441.1 DS231818 EDS41347.1 AXCN02001805 AAAB01008979 EAA13769.4 APCN01003485 GANO01000560 JAB59311.1 CH477188 EAT48811.1 JXUM01001263 JXUM01001264 KQ560119 KXJ84380.1 ATLV01019337 KE525268 KFB44053.1 AXCM01006588 GGFK01003502 MBW36823.1 GGFJ01000109 MBW49250.1 GGFJ01000108 MBW49249.1 JH431806 GGFK01003134 MBW36455.1 GGFJ01000110 MBW49251.1 ADMH02001311 ETN63054.1 ABJB010908985 DS958725 EEC19290.1 GEFM01006110 JAP69686.1 GANP01002762 JAB81706.1 GFWV01013093 MAA37822.1 MWRG01004097 PRD30826.1 GANP01014563 JAB69905.1 GBXI01007327 JAD06965.1 GBXI01014386 JAC99905.1 GFWV01005852 MAA30582.1 NNAY01002664 OXU20832.1 JXJN01021533 JXJN01021534 GAMC01008352 GAMC01008349 JAB98203.1 CCAG010016028 GACK01009462 JAA55572.1 GEDV01004284 JAP84273.1 GDHF01002387 JAI49927.1 GDHF01032642 JAI19672.1 GFAC01002912 JAT96276.1 JRES01001143 KNC25163.1 GAMC01008354 GAMC01008351 JAB98204.1 GAKP01021073 JAC37879.1 GEFH01002084 JAP66497.1 CP012528 ALC49915.1 GFPF01010546 MAA21692.1 CH964239 EDW82565.1 CH933815 EDW07927.1 GFAA01002135 JAU01300.1 KRG07526.1 CH379063 KRT06173.1 CVRI01000040 CRK94872.1
Proteomes
UP000007151
UP000005204
UP000283053
UP000027135
UP000009046
UP000007266
+ More
UP000218220 UP000007819 UP000235965 UP000000311 UP000036403 UP000030742 UP000005203 UP000242457 UP000078540 UP000008237 UP000005205 UP000078541 UP000078542 UP000078492 UP000053097 UP000279307 UP000075884 UP000002320 UP000075886 UP000075885 UP000075902 UP000007062 UP000075900 UP000075840 UP000075880 UP000075920 UP000075881 UP000008820 UP000069940 UP000249989 UP000076408 UP000030765 UP000075883 UP000069272 UP000000673 UP000001555 UP000076407 UP000095300 UP000078200 UP000215335 UP000092443 UP000092460 UP000091820 UP000092445 UP000092444 UP000002358 UP000037069 UP000095301 UP000092553 UP000007798 UP000009192 UP000001819 UP000183832
UP000218220 UP000007819 UP000235965 UP000000311 UP000036403 UP000030742 UP000005203 UP000242457 UP000078540 UP000008237 UP000005205 UP000078541 UP000078542 UP000078492 UP000053097 UP000279307 UP000075884 UP000002320 UP000075886 UP000075885 UP000075902 UP000007062 UP000075900 UP000075840 UP000075880 UP000075920 UP000075881 UP000008820 UP000069940 UP000249989 UP000076408 UP000030765 UP000075883 UP000069272 UP000000673 UP000001555 UP000076407 UP000095300 UP000078200 UP000215335 UP000092443 UP000092460 UP000091820 UP000092445 UP000092444 UP000002358 UP000037069 UP000095301 UP000092553 UP000007798 UP000009192 UP000001819 UP000183832
PRIDE
Pfam
Interpro
IPR015433
PI_Kinase
+ More
IPR016024 ARM-type_fold
IPR001263 PI3K_accessory_dom
IPR036940 PI3/4_kinase_cat_sf
IPR011009 Kinase-like_dom_sf
IPR000403 PI3/4_kinase_cat_dom
IPR018936 PI3/4_kinase_CS
IPR042236 PI3K_accessory_sf
IPR001267 Thymidine_kinase
IPR027417 P-loop_NTPase
IPR020633 Thymidine_kinase_CS
IPR007330 MIT
IPR022684 Calpain_cysteine_protease
IPR038765 Papain-like_cys_pep_sf
IPR022683 Calpain_III
IPR001300 Peptidase_C2_calpain_cat
IPR036181 MIT_dom_sf
IPR022682 Calpain_domain_III
IPR036213 Calpain_III_sf
IPR016024 ARM-type_fold
IPR001263 PI3K_accessory_dom
IPR036940 PI3/4_kinase_cat_sf
IPR011009 Kinase-like_dom_sf
IPR000403 PI3/4_kinase_cat_dom
IPR018936 PI3/4_kinase_CS
IPR042236 PI3K_accessory_sf
IPR001267 Thymidine_kinase
IPR027417 P-loop_NTPase
IPR020633 Thymidine_kinase_CS
IPR007330 MIT
IPR022684 Calpain_cysteine_protease
IPR038765 Papain-like_cys_pep_sf
IPR022683 Calpain_III
IPR001300 Peptidase_C2_calpain_cat
IPR036181 MIT_dom_sf
IPR022682 Calpain_domain_III
IPR036213 Calpain_III_sf
SUPFAM
Gene 3D
CDD
ProteinModelPortal
A0A212FME8
H9JTG8
A0A3S2TCX0
A0A067QIG5
E0W3P8
A0A069DZT9
+ More
A0A023F555 A0A1B6HW70 A0A1B6LF78 A0A0T6BCD2 D2A3W3 A0A2A4JVW0 A0A0A9ZGW4 A0A0A9ZFG5 A0A2H8TZ06 A0A2S2Q5K8 J9JMX5 A0A2J7QZA3 A0A1Y1M0R4 A0A0C9QK07 E2AJ31 A0A0J7L641 U4U5D1 A0A088A932 A0A2A3EB47 A0A195AUW7 E2B3H5 A0A158NNB0 A0A195FY16 A0A195CS11 A0A151IT87 A0A026WVJ6 A0A3L8DIY4 A0A1Q3G3S1 A0A182NV87 B0W0P5 A0A182QGM5 A0A182P1W5 A0A182TSY5 Q7Q237 A0A182RUL6 A0A182I5Q9 W4VRQ0 A0A182JI18 A0A182W9I8 A0A182JW38 A0A1S4EV63 Q17PY3 A0A182GH84 A0A182YIY1 A0A084W1F9 A0A1S4EV62 A0A182MWN4 A0A2M4A7Q3 A0A2M4B8D4 A0A2M4B873 T1J2J9 A0A2M4A6S0 A0A182FVH4 A0A2M4B8D6 W5JJV1 B7QKB8 A0A131XVJ2 V5IIQ8 A0A293MAF0 A0A182X0H0 A0A1I8Q522 A0A1I8Q510 A0A2P6KWY6 V5GI35 A0A0A1X897 A0A0A1WMM2 A0A293LPG1 A0A1A9ULE3 A0A232ER76 A0A1A9XE14 A0A1B0BVY8 A0A1A9W400 A0A1B0AC15 W8BMJ7 A0A1B0FEG5 L7LVG2 A0A131YYL1 K7J6G1 A0A0K8WFR5 A0A0K8TYY2 A0A1E1XB13 A0A0L0BYP1 W8C6T2 A0A034V5U8 A0A1I8M8W2 A0A131XIY6 A0A0M4ETM9 A0A224Z5V5 B4NCK6 B4L8F2 A0A1E1XQJ6 A0A0Q9XGA2 A0A0R3P3H5 A0A1J1I3I5
A0A023F555 A0A1B6HW70 A0A1B6LF78 A0A0T6BCD2 D2A3W3 A0A2A4JVW0 A0A0A9ZGW4 A0A0A9ZFG5 A0A2H8TZ06 A0A2S2Q5K8 J9JMX5 A0A2J7QZA3 A0A1Y1M0R4 A0A0C9QK07 E2AJ31 A0A0J7L641 U4U5D1 A0A088A932 A0A2A3EB47 A0A195AUW7 E2B3H5 A0A158NNB0 A0A195FY16 A0A195CS11 A0A151IT87 A0A026WVJ6 A0A3L8DIY4 A0A1Q3G3S1 A0A182NV87 B0W0P5 A0A182QGM5 A0A182P1W5 A0A182TSY5 Q7Q237 A0A182RUL6 A0A182I5Q9 W4VRQ0 A0A182JI18 A0A182W9I8 A0A182JW38 A0A1S4EV63 Q17PY3 A0A182GH84 A0A182YIY1 A0A084W1F9 A0A1S4EV62 A0A182MWN4 A0A2M4A7Q3 A0A2M4B8D4 A0A2M4B873 T1J2J9 A0A2M4A6S0 A0A182FVH4 A0A2M4B8D6 W5JJV1 B7QKB8 A0A131XVJ2 V5IIQ8 A0A293MAF0 A0A182X0H0 A0A1I8Q522 A0A1I8Q510 A0A2P6KWY6 V5GI35 A0A0A1X897 A0A0A1WMM2 A0A293LPG1 A0A1A9ULE3 A0A232ER76 A0A1A9XE14 A0A1B0BVY8 A0A1A9W400 A0A1B0AC15 W8BMJ7 A0A1B0FEG5 L7LVG2 A0A131YYL1 K7J6G1 A0A0K8WFR5 A0A0K8TYY2 A0A1E1XB13 A0A0L0BYP1 W8C6T2 A0A034V5U8 A0A1I8M8W2 A0A131XIY6 A0A0M4ETM9 A0A224Z5V5 B4NCK6 B4L8F2 A0A1E1XQJ6 A0A0Q9XGA2 A0A0R3P3H5 A0A1J1I3I5
PDB
6BQ1
E-value=0,
Score=2478
Ontologies
PATHWAY
GO
PANTHER
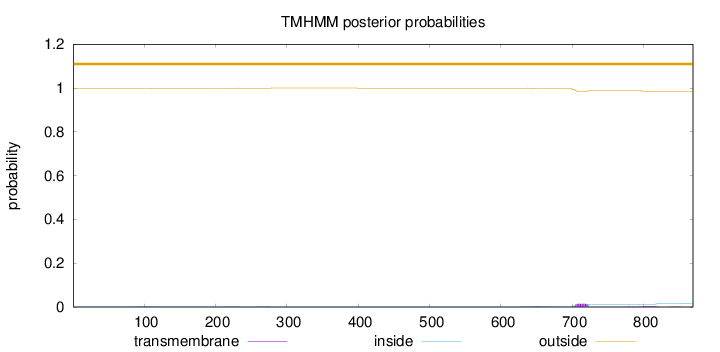
Topology
Length:
869
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.4342
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00221
outside
1 - 869
Population Genetic Test Statistics
Pi
154.091784
Theta
160.935761
Tajima's D
0.627721
CLR
0.079978
CSRT
0.549622518874056
Interpretation
Uncertain
