Gene
KWMTBOMO09553 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013021
Annotation
fructose_1?6-bisphosphate_aldolase_[Bombyx_mori]
Full name
Fructose-bisphosphate aldolase
Location in the cell
Cytoplasmic Reliability : 1.363 Mitochondrial Reliability : 1.577
Sequence
CDS
ATGTCCACCTACTTTCAATACCCAACTCCTGAGCTGCAGGAGGAGCTCAAGAAGATCGCTCAAGCAATTGTAGCTCCCGCAAAGGGTATCCTGGCCGCTGACGAATCCACCGGTACAATGGGCAAGCGTTTGCAGGACATCGGCGTGGAGAACACAGAGGAGAACCGTCGTCGTTATCGCCAACTGCTATTCAGCTCTGACGCTGTGCTCTCCGAGAACATCTCTGGTGTGATCCTGTTCCACGAGACCCTTTACCAGAAGGCTGACGATGGAACCCCTCTGGTCTCCCTGCTGGAGAAGAAGGGCATCATCCCCGGCATCAAGGTCGACAAGGGTGTCGTCCCGCTGTTCGGATCGGAAGACGAATGCACCACCCAGGGTCTGGACGACCTCGCCCAGCGCTGCGCCCAGTACAAGAAGGACGGCTGCCACTTCGCCAAGTGGCGTTGCGTGCTGAAGATTGGCCGCAACACCCCCTCGTACCAAGCTATCCAGGAAAACGCCAATGTTCTCGCCCGCTACGCCTCCATCTGTCAGAGCCAACGCATCGTGCCAATTGTCGAGCCCGAAGTCTTACCTGATGGCGAGCACGACCTGGACCGCGCCCAGAAGGTGACTGAGGTCGTTCTGGCCGCCGTGTATAAGGCGCTGAACGACCACCACGTGTACCTCGAGGGGACACTCCTCAAGCCCAACATGGTGACGGCCGGTCAGTCGTGCAAGAAGACGTACACTCCCAACGACGTCGCGCGCGCCACCGTCACCGCCCTCCTCCGCACCGTGCCCGCCGCAGTCCCCGGAGTGACATTCCTCTCCGGTGGTCAGTCCGAAGAGGAGGCCTCGGTGCATCTGAACGCTATCAACGCTGTGGACCTCAAGAGGCCCTGGGTTCTGACGTTCAGCTACGGGCGCGCCCTCCAGGCTTCGGTGCTCCGCGCCTGGGGAGGGAAGACCGAGAACATTCTCGCCGGACAGCAGGAGCTGATCAAGCGTGCTAAGGCCAACGGTCTCGCAGCAGTCGGCAAATACGTTGCCGGCTCTATTCCTTCGCTGGCCGCTTCTAAATCGAACTTTGTCAAATCTCACGCTTATTAA
Protein
MSTYFQYPTPELQEELKKIAQAIVAPAKGILAADESTGTMGKRLQDIGVENTEENRRRYRQLLFSSDAVLSENISGVILFHETLYQKADDGTPLVSLLEKKGIIPGIKVDKGVVPLFGSEDECTTQGLDDLAQRCAQYKKDGCHFAKWRCVLKIGRNTPSYQAIQENANVLARYASICQSQRIVPIVEPEVLPDGEHDLDRAQKVTEVVLAAVYKALNDHHVYLEGTLLKPNMVTAGQSCKKTYTPNDVARATVTALLRTVPAAVPGVTFLSGGQSEEEASVHLNAINAVDLKRPWVLTFSYGRALQASVLRAWGGKTENILAGQQELIKRAKANGLAAVGKYVAGSIPSLAASKSNFVKSHAY
Summary
Description
May take part in developmental stage-specific or tissue -specific sugar-phosphate metabolisms. Protein acts on two substrates fructose 1,6-bisphosphate and fructose 1-phosphate (like other class I aldolases).
Catalytic Activity
beta-D-fructose 1,6-bisphosphate = D-glyceraldehyde 3-phosphate + dihydroxyacetone phosphate
Biophysicochemical Properties
Optimum pH is 7.2-7.8 for isozyme alpha, and 7.2-7.5 for isozymes beta and gamma.
Subunit
Homotetramer.
Similarity
Belongs to the class I fructose-bisphosphate aldolase family.
Keywords
3D-structure
Acetylation
Alternative splicing
Complete proteome
Direct protein sequencing
Glycolysis
Lyase
Reference proteome
Schiff base
Feature
chain Fructose-bisphosphate aldolase
splice variant In isoform Beta.
splice variant In isoform Beta.
Uniprot
Q1HPN7
Q75PQ3
A0A1E1W008
S4PKR2
A0A194R6Y9
A0A2A4K0C6
+ More
A0A1E1WFZ0 A0A194Q080 I4DS64 I4DIX5 A0A2W1BGR3 A0A437BC03 A0A212EN62 A0A222AJC5 Q6PPI0 A0A0A1WLY1 Q5XXS8 A0A034W0D2 A0A0C9RLK5 A0A1I8MI51 T1PF08 W8BB97 A0A034VYR0 A0A1L8EHG5 A0A1I8MI52 A0A0A1XBW0 A0A1I8Q820 A0A0L0CB25 A0A2J7Q6X2 A0A1L8EEB3 A0A1L8EEC0 A0A0Q9WWZ4 A0A2J7Q6X5 A0A2P0XIZ4 G9C5D5 A0A1B0CFJ1 A0A3B0JS88 A0A1L8E1Y6 A0A0P8Y2A2 A0A0Q9WQB4 A0A067QX08 A0A3B0KE28 D3TPU5 A0A0Q9XD95 A0A1W4VKK0 A0A1A9WAA9 A0A1I8JU48 A0A182YQJ6 A0A1I8JV83 Q178U8 A0A1B6E5I5 A0A1I8JV79 A0A182P065 E0W3R5 A0A182H5C4 A0A182Q2N0 E9GJ13 A0A1Y9IY90 U5EXS9 A0A182TX64 A0A182UVD5 Q7PGI9 A0A182HSP3 A0A1A9ULU3 A0A2M3ZZ82 A0A182JDC8 A0A2M3YXN1 A0A182FNW4 A0A182K296 A0A182LER9 A0A1Y1MII1 A0A068FCL9 A0A182MF90 A0A1Q3G4B3 A0A182VWX4 A0A0P6HXA7 A0A0M4ENX9 W5JKF9 P07764-3 B4NKQ7 F3YDE2 A0A3R7PR20 B4ICK7 B4QW40 A0A226E871 A0A0P5MZC0 A0A1B0GEZ8 F3YDB5 A0A2R7X4P6 A0A3B0JQQ2 A0A0P4WTD2 Q297A9 A0A0P5G3E3 D3TPV3 A0A0P5Y3V4 A0A1A9ZP50 P07764-2 A4V3G1 A0A0P5Y3V0 A5XCY1
A0A1E1WFZ0 A0A194Q080 I4DS64 I4DIX5 A0A2W1BGR3 A0A437BC03 A0A212EN62 A0A222AJC5 Q6PPI0 A0A0A1WLY1 Q5XXS8 A0A034W0D2 A0A0C9RLK5 A0A1I8MI51 T1PF08 W8BB97 A0A034VYR0 A0A1L8EHG5 A0A1I8MI52 A0A0A1XBW0 A0A1I8Q820 A0A0L0CB25 A0A2J7Q6X2 A0A1L8EEB3 A0A1L8EEC0 A0A0Q9WWZ4 A0A2J7Q6X5 A0A2P0XIZ4 G9C5D5 A0A1B0CFJ1 A0A3B0JS88 A0A1L8E1Y6 A0A0P8Y2A2 A0A0Q9WQB4 A0A067QX08 A0A3B0KE28 D3TPU5 A0A0Q9XD95 A0A1W4VKK0 A0A1A9WAA9 A0A1I8JU48 A0A182YQJ6 A0A1I8JV83 Q178U8 A0A1B6E5I5 A0A1I8JV79 A0A182P065 E0W3R5 A0A182H5C4 A0A182Q2N0 E9GJ13 A0A1Y9IY90 U5EXS9 A0A182TX64 A0A182UVD5 Q7PGI9 A0A182HSP3 A0A1A9ULU3 A0A2M3ZZ82 A0A182JDC8 A0A2M3YXN1 A0A182FNW4 A0A182K296 A0A182LER9 A0A1Y1MII1 A0A068FCL9 A0A182MF90 A0A1Q3G4B3 A0A182VWX4 A0A0P6HXA7 A0A0M4ENX9 W5JKF9 P07764-3 B4NKQ7 F3YDE2 A0A3R7PR20 B4ICK7 B4QW40 A0A226E871 A0A0P5MZC0 A0A1B0GEZ8 F3YDB5 A0A2R7X4P6 A0A3B0JQQ2 A0A0P4WTD2 Q297A9 A0A0P5G3E3 D3TPV3 A0A0P5Y3V4 A0A1A9ZP50 P07764-2 A4V3G1 A0A0P5Y3V0 A5XCY1
EC Number
4.1.2.13
Pubmed
19121390
23622113
26354079
22651552
28756777
22118469
+ More
25830018 25348373 25315136 24495485 26108605 17994087 21835214 18057021 24845553 20353571 25244985 17510324 20566863 26483478 21292972 12364791 14747013 17210077 20966253 28004739 20920257 23761445 1339430 1740444 1732743 7487099 17379620 10731132 12537572 12537569 3918528 3140728 6805442 8537310 1959612 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085
25830018 25348373 25315136 24495485 26108605 17994087 21835214 18057021 24845553 20353571 25244985 17510324 20566863 26483478 21292972 12364791 14747013 17210077 20966253 28004739 20920257 23761445 1339430 1740444 1732743 7487099 17379620 10731132 12537572 12537569 3918528 3140728 6805442 8537310 1959612 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085
EMBL
BABH01003873
BABH01003874
BABH01003875
DQ443365
LC316985
ABF51454.1
+ More
BBA26324.1 AB166792 BAD12426.1 GDQN01010734 JAT80320.1 GAIX01004365 JAA88195.1 KQ460878 KPJ11631.1 NWSH01000318 PCG77509.1 GDQN01005165 JAT85889.1 KQ459582 KPI98987.1 AK405438 BAM20754.1 AK401243 BAM17865.1 KZ150073 PZC74008.1 RSAL01000093 RVE47930.1 AGBW02013713 OWR42930.1 KY609709 ASO76464.1 AY588066 AAT01078.1 GBXI01014752 JAC99539.1 AY725784 AAU95197.1 GAKP01011764 JAC47188.1 GBYB01007841 JAG77608.1 KA647331 AFP61960.1 GAMC01016134 JAB90421.1 GAKP01011765 JAC47187.1 GFDG01000766 JAV18033.1 GBXI01016995 GBXI01005856 JAC97296.1 JAD08436.1 JRES01000664 KNC29432.1 NEVH01017450 PNF24336.1 GFDG01001747 JAV17052.1 GFDG01001742 JAV17057.1 CH964272 KRF99976.1 PNF24337.1 KY661328 AVA17415.1 HQ851376 AEV89754.1 AJWK01010111 AJWK01010112 AJWK01010113 AJWK01010114 OUUW01000007 SPP83282.1 GFDF01001353 JAV12731.1 CH902617 KPU80854.1 CH940650 KRF83012.1 KK853259 KDR09236.1 SPP83281.1 EZ423447 ADD19723.1 CH933806 KRG01699.1 CH477359 EAT42703.1 GEDC01009708 GEDC01004098 JAS27590.1 JAS33200.1 DS235882 EEB20271.1 JXUM01025903 JXUM01025904 KQ560737 KXJ81075.1 AXCN02000003 GL732547 EFX80562.1 GANO01000784 JAB59087.1 AAAB01008859 EAA44916.4 APCN01000316 GGFK01000449 MBW33770.1 GGFM01000272 MBW21023.1 GEZM01032210 JAV84768.1 KJ686589 MH675996 AID61753.1 QBO59887.1 AXCM01006740 GFDL01000393 JAV34652.1 GDIP01252032 GDIP01169946 GDIP01095311 GDIQ01037424 JAI71369.1 JAN57313.1 CP012526 ALC45697.1 ADMH02000939 ETN64621.1 D10446 M98351 X60064 D10762 DQ864133 DQ864134 DQ864135 DQ864136 DQ864137 DQ864138 DQ864139 DQ864140 DQ864141 DQ864142 DQ864143 DQ864144 AE014297 AY058667 AY128452 EDW84118.2 BT126303 AEB39664.1 AGB96365.1 QCYY01001906 ROT74380.1 CH480828 EDW45103.1 CM000364 EDX14539.1 LNIX01000006 OXA52856.1 GDIQ01160575 JAK91150.1 CCAG010012689 BT126276 AEB39637.1 KK856759 PTY26380.1 SPP83283.1 GDIP01251088 JAI72313.1 CM000070 EAL28297.4 GDIQ01251700 JAK00025.1 EZ423455 ADD19731.1 GDIP01063363 JAM40352.1 BT126163 AAN14383.3 ADZ99415.1 GDIP01063362 JAM40353.1 DQ864132 ABH06767.1
BBA26324.1 AB166792 BAD12426.1 GDQN01010734 JAT80320.1 GAIX01004365 JAA88195.1 KQ460878 KPJ11631.1 NWSH01000318 PCG77509.1 GDQN01005165 JAT85889.1 KQ459582 KPI98987.1 AK405438 BAM20754.1 AK401243 BAM17865.1 KZ150073 PZC74008.1 RSAL01000093 RVE47930.1 AGBW02013713 OWR42930.1 KY609709 ASO76464.1 AY588066 AAT01078.1 GBXI01014752 JAC99539.1 AY725784 AAU95197.1 GAKP01011764 JAC47188.1 GBYB01007841 JAG77608.1 KA647331 AFP61960.1 GAMC01016134 JAB90421.1 GAKP01011765 JAC47187.1 GFDG01000766 JAV18033.1 GBXI01016995 GBXI01005856 JAC97296.1 JAD08436.1 JRES01000664 KNC29432.1 NEVH01017450 PNF24336.1 GFDG01001747 JAV17052.1 GFDG01001742 JAV17057.1 CH964272 KRF99976.1 PNF24337.1 KY661328 AVA17415.1 HQ851376 AEV89754.1 AJWK01010111 AJWK01010112 AJWK01010113 AJWK01010114 OUUW01000007 SPP83282.1 GFDF01001353 JAV12731.1 CH902617 KPU80854.1 CH940650 KRF83012.1 KK853259 KDR09236.1 SPP83281.1 EZ423447 ADD19723.1 CH933806 KRG01699.1 CH477359 EAT42703.1 GEDC01009708 GEDC01004098 JAS27590.1 JAS33200.1 DS235882 EEB20271.1 JXUM01025903 JXUM01025904 KQ560737 KXJ81075.1 AXCN02000003 GL732547 EFX80562.1 GANO01000784 JAB59087.1 AAAB01008859 EAA44916.4 APCN01000316 GGFK01000449 MBW33770.1 GGFM01000272 MBW21023.1 GEZM01032210 JAV84768.1 KJ686589 MH675996 AID61753.1 QBO59887.1 AXCM01006740 GFDL01000393 JAV34652.1 GDIP01252032 GDIP01169946 GDIP01095311 GDIQ01037424 JAI71369.1 JAN57313.1 CP012526 ALC45697.1 ADMH02000939 ETN64621.1 D10446 M98351 X60064 D10762 DQ864133 DQ864134 DQ864135 DQ864136 DQ864137 DQ864138 DQ864139 DQ864140 DQ864141 DQ864142 DQ864143 DQ864144 AE014297 AY058667 AY128452 EDW84118.2 BT126303 AEB39664.1 AGB96365.1 QCYY01001906 ROT74380.1 CH480828 EDW45103.1 CM000364 EDX14539.1 LNIX01000006 OXA52856.1 GDIQ01160575 JAK91150.1 CCAG010012689 BT126276 AEB39637.1 KK856759 PTY26380.1 SPP83283.1 GDIP01251088 JAI72313.1 CM000070 EAL28297.4 GDIQ01251700 JAK00025.1 EZ423455 ADD19731.1 GDIP01063363 JAM40352.1 BT126163 AAN14383.3 ADZ99415.1 GDIP01063362 JAM40353.1 DQ864132 ABH06767.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000053268
UP000283053
UP000007151
+ More
UP000095301 UP000095300 UP000037069 UP000235965 UP000007798 UP000092461 UP000268350 UP000007801 UP000008792 UP000027135 UP000009192 UP000192221 UP000091820 UP000075900 UP000076408 UP000075903 UP000008820 UP000075885 UP000009046 UP000069940 UP000249989 UP000075886 UP000000305 UP000076407 UP000075902 UP000007062 UP000075840 UP000078200 UP000075880 UP000069272 UP000075881 UP000075882 UP000075883 UP000075920 UP000092553 UP000000673 UP000000803 UP000283509 UP000001292 UP000000304 UP000198287 UP000092444 UP000001819 UP000092445
UP000095301 UP000095300 UP000037069 UP000235965 UP000007798 UP000092461 UP000268350 UP000007801 UP000008792 UP000027135 UP000009192 UP000192221 UP000091820 UP000075900 UP000076408 UP000075903 UP000008820 UP000075885 UP000009046 UP000069940 UP000249989 UP000075886 UP000000305 UP000076407 UP000075902 UP000007062 UP000075840 UP000078200 UP000075880 UP000069272 UP000075881 UP000075882 UP000075883 UP000075920 UP000092553 UP000000673 UP000000803 UP000283509 UP000001292 UP000000304 UP000198287 UP000092444 UP000001819 UP000092445
Pfam
PF00274 Glycolytic
Gene 3D
ProteinModelPortal
Q1HPN7
Q75PQ3
A0A1E1W008
S4PKR2
A0A194R6Y9
A0A2A4K0C6
+ More
A0A1E1WFZ0 A0A194Q080 I4DS64 I4DIX5 A0A2W1BGR3 A0A437BC03 A0A212EN62 A0A222AJC5 Q6PPI0 A0A0A1WLY1 Q5XXS8 A0A034W0D2 A0A0C9RLK5 A0A1I8MI51 T1PF08 W8BB97 A0A034VYR0 A0A1L8EHG5 A0A1I8MI52 A0A0A1XBW0 A0A1I8Q820 A0A0L0CB25 A0A2J7Q6X2 A0A1L8EEB3 A0A1L8EEC0 A0A0Q9WWZ4 A0A2J7Q6X5 A0A2P0XIZ4 G9C5D5 A0A1B0CFJ1 A0A3B0JS88 A0A1L8E1Y6 A0A0P8Y2A2 A0A0Q9WQB4 A0A067QX08 A0A3B0KE28 D3TPU5 A0A0Q9XD95 A0A1W4VKK0 A0A1A9WAA9 A0A1I8JU48 A0A182YQJ6 A0A1I8JV83 Q178U8 A0A1B6E5I5 A0A1I8JV79 A0A182P065 E0W3R5 A0A182H5C4 A0A182Q2N0 E9GJ13 A0A1Y9IY90 U5EXS9 A0A182TX64 A0A182UVD5 Q7PGI9 A0A182HSP3 A0A1A9ULU3 A0A2M3ZZ82 A0A182JDC8 A0A2M3YXN1 A0A182FNW4 A0A182K296 A0A182LER9 A0A1Y1MII1 A0A068FCL9 A0A182MF90 A0A1Q3G4B3 A0A182VWX4 A0A0P6HXA7 A0A0M4ENX9 W5JKF9 P07764-3 B4NKQ7 F3YDE2 A0A3R7PR20 B4ICK7 B4QW40 A0A226E871 A0A0P5MZC0 A0A1B0GEZ8 F3YDB5 A0A2R7X4P6 A0A3B0JQQ2 A0A0P4WTD2 Q297A9 A0A0P5G3E3 D3TPV3 A0A0P5Y3V4 A0A1A9ZP50 P07764-2 A4V3G1 A0A0P5Y3V0 A5XCY1
A0A1E1WFZ0 A0A194Q080 I4DS64 I4DIX5 A0A2W1BGR3 A0A437BC03 A0A212EN62 A0A222AJC5 Q6PPI0 A0A0A1WLY1 Q5XXS8 A0A034W0D2 A0A0C9RLK5 A0A1I8MI51 T1PF08 W8BB97 A0A034VYR0 A0A1L8EHG5 A0A1I8MI52 A0A0A1XBW0 A0A1I8Q820 A0A0L0CB25 A0A2J7Q6X2 A0A1L8EEB3 A0A1L8EEC0 A0A0Q9WWZ4 A0A2J7Q6X5 A0A2P0XIZ4 G9C5D5 A0A1B0CFJ1 A0A3B0JS88 A0A1L8E1Y6 A0A0P8Y2A2 A0A0Q9WQB4 A0A067QX08 A0A3B0KE28 D3TPU5 A0A0Q9XD95 A0A1W4VKK0 A0A1A9WAA9 A0A1I8JU48 A0A182YQJ6 A0A1I8JV83 Q178U8 A0A1B6E5I5 A0A1I8JV79 A0A182P065 E0W3R5 A0A182H5C4 A0A182Q2N0 E9GJ13 A0A1Y9IY90 U5EXS9 A0A182TX64 A0A182UVD5 Q7PGI9 A0A182HSP3 A0A1A9ULU3 A0A2M3ZZ82 A0A182JDC8 A0A2M3YXN1 A0A182FNW4 A0A182K296 A0A182LER9 A0A1Y1MII1 A0A068FCL9 A0A182MF90 A0A1Q3G4B3 A0A182VWX4 A0A0P6HXA7 A0A0M4ENX9 W5JKF9 P07764-3 B4NKQ7 F3YDE2 A0A3R7PR20 B4ICK7 B4QW40 A0A226E871 A0A0P5MZC0 A0A1B0GEZ8 F3YDB5 A0A2R7X4P6 A0A3B0JQQ2 A0A0P4WTD2 Q297A9 A0A0P5G3E3 D3TPV3 A0A0P5Y3V4 A0A1A9ZP50 P07764-2 A4V3G1 A0A0P5Y3V0 A5XCY1
PDB
1FBA
E-value=4.71209e-162,
Score=1465
Ontologies
PATHWAY
00010
Glycolysis / Gluconeogenesis - Bombyx mori (domestic silkworm)
00030 Pentose phosphate pathway - Bombyx mori (domestic silkworm)
00051 Fructose and mannose metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
00030 Pentose phosphate pathway - Bombyx mori (domestic silkworm)
00051 Fructose and mannose metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
GO
Topology
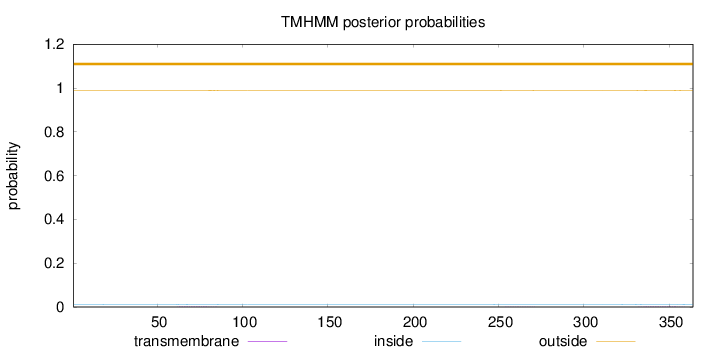
Length:
364
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.08109
Exp number, first 60 AAs:
0.00024
Total prob of N-in:
0.01222
outside
1 - 364
Population Genetic Test Statistics
Pi
224.458604
Theta
176.254075
Tajima's D
1.134993
CLR
0.201982
CSRT
0.698315084245788
Interpretation
Uncertain
