Gene
KWMTBOMO09551
Pre Gene Modal
BGIBMGA012826
Annotation
PREDICTED:_uncharacterized_protein_LOC106129912_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 3.514
Sequence
CDS
ATGGAACAGTCGACGCTCATGTCTCCGGGACAAGCTCTGATGGGGATGGACGCGAAGGATCGACTCGTGGAGGAGATATTACTCCTAGCGCTGTTCCTTGTGTCGACGGCGGCAAACTCGCTCGCCCTACTCGTTTTCTGCAGGCGACCCGGTCTCAGAACTATATCGAATCGGTTCGTCATCAATCTTCTGGCGACGAATCTATTGACTACTTCATTGTTGGCGCCCGCGCTCTTCGGCGAGCATACGACGAAGAAAAACGCCAGTCTGTGGGTGGAGGCAGGCGACTCGGCGGCCTCAGCCTGCAGCCTGTCAGCCCTGTTGTCCGTAATGCTGATTTCACTCGACCAGCACCACGCCGTCACGGATCCTCTTCGCTGCCACACCCGTCTCTTACAGCGTCGTCCAAGCTCGCTCTGCGCCGCCACCTGGGTCGCCGCGGCATCCGCAGCCGCAATTCGAGCTACCATCGCCGTGTTCCACTACTACTATCCGACCGAATCAGCGCTGCAAGGTATCCAGCTCGCCTATTACGTCGTGTACTTGATATTGGGAGTGATCGCACCGATAACTATAGTTTGCGTCATGTACGCGAGAATCTACCGTGCGGCGCGCTCGTCTGGATTACGAGCTCGTGCGAACGGTGCCAGTGCTCTGCAGTTACACGAGCCCCCGCCCCCCCTTCCGAACCTCATCGAAGAGGGGGAAGGAGAGGGACTCACACTTAGAATAGACGGATCAGAAGACGTAGCGTCCGGAGAGCACGATCGCAACAAATTTGGACCGTTTCTGTTACCACCGTCAGAGAGGCCAGTCCGTTGCCCATCTGTTGCCAGTCTACGGAATTCCCGCACTAAAGATGGAAAGTCGAACGAAGATCTGAGGAAGGGATTACCGTCTGTTAGTTCGGCACCATCACTCGCTGCCCCTCTCTTGGCGGTGCAGCAACGGACCCCTCAGACCTCACACAATGGATCTAGAATAACATCGATACGATGCCGCATTTCAAACGCCTCCCTGTTCCGTTATAGAGAAGAATCTCGTGCGGCTCGCGTCGGTTTGCTGACGGGGGCGTTGGCCATGCTGCACGTCCCGCATGCCGCAGCCGCCGTGGCCTCCGCCGCTGTCCCCGGGCTGGCTGATTGTGCGCGCACCGCATCGCTCGCGTTACTCCTGCTTGCCTCTGCGGCTTCACCGTTCCTGTTTGCACTACGTTCGCGCCGGGTGCAGCGTGAGGCTCGGCGTCTGTTCGTGCCTGAGAAGAGCGCTTGGGAGAGAGCATCGGCGTCCGCGTCGTCTGCGGCGGCGGACGGGCAACGCGCTAAGGCCGCTTTGGAGTTGCTGCGGACATTGTCGCCGAGCAGCGGAGAGCCAGCGCCGGACAGCTCGGCGGAGTCGGGTCCTTGCCGCGGCTCGTTCAGCTCTGGGGGCAGCACGCAGCTCTCGTCAGCTTCGGACGAGTGA
Protein
MEQSTLMSPGQALMGMDAKDRLVEEILLLALFLVSTAANSLALLVFCRRPGLRTISNRFVINLLATNLLTTSLLAPALFGEHTTKKNASLWVEAGDSAASACSLSALLSVMLISLDQHHAVTDPLRCHTRLLQRRPSSLCAATWVAAASAAAIRATIAVFHYYYPTESALQGIQLAYYVVYLILGVIAPITIVCVMYARIYRAARSSGLRARANGASALQLHEPPPPLPNLIEEGEGEGLTLRIDGSEDVASGEHDRNKFGPFLLPPSERPVRCPSVASLRNSRTKDGKSNEDLRKGLPSVSSAPSLAAPLLAVQQRTPQTSHNGSRITSIRCRISNASLFRYREESRAARVGLLTGALAMLHVPHAAAAVASAAVPGLADCARTASLALLLLASAASPFLFALRSRRVQREARRLFVPEKSAWERASASASSAAADGQRAKAALELLRTLSPSSGEPAPDSSAESGPCRGSFSSGGSTQLSSASDE
Summary
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF00001 7tm_1
ProteinModelPortal
Ontologies
GO
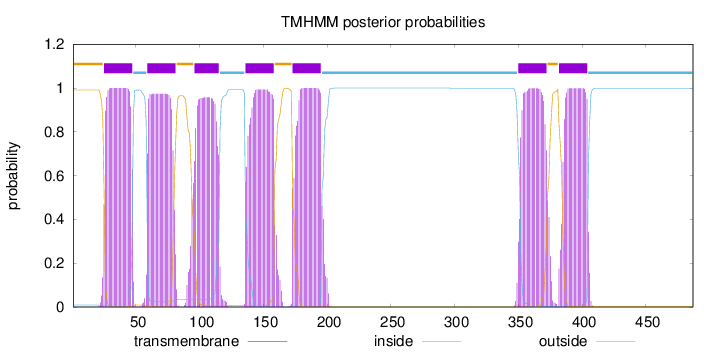
Topology
Length:
487
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
151.49017
Exp number, first 60 AAs:
24.03938
Total prob of N-in:
0.00883
POSSIBLE N-term signal
sequence
outside
1 - 24
TMhelix
25 - 47
inside
48 - 58
TMhelix
59 - 81
outside
82 - 95
TMhelix
96 - 115
inside
116 - 135
TMhelix
136 - 158
outside
159 - 172
TMhelix
173 - 195
inside
196 - 349
TMhelix
350 - 372
outside
373 - 381
TMhelix
382 - 404
inside
405 - 487
Population Genetic Test Statistics
Pi
212.150129
Theta
164.643189
Tajima's D
1.124369
CLR
0.215674
CSRT
0.699315034248288
Interpretation
Uncertain
