Gene
KWMTBOMO09549
Pre Gene Modal
BGIBMGA012823
Annotation
PREDICTED:_uncharacterized_protein_LOC101743839_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.381
Sequence
CDS
ATGTCACTGAAAGCTTACGAAGACGCCTGCTTAAGAGCGGAGCTGTTCGGTCAGCCTTTACCGGACAAAGATGAATTTATCAGGAAAAATAAACATTTAGATATTGTTGAATTCGAAGAAGTGGAAATAAAAACTGCTGAGAATACGGCTATGCTTCATGATCAAATGAAGGAAGGCACGACTGGTCTCACGGAACTCAACACTATCCTAGATTCAACTCAAACGAAAATAAATCGACTAAAAGGCGTTTGCGGTAGCATCACGAATTTCTTCCGAGTCAAATTGAATTCCAAAGATAATATTTCGTATTCTAGCGACCCCAGTTACGTAGGTCAAACGAATTTCCAAAGAGACTACGTGCCGGTTTCGAACCGAATCGACTCTGTGAACGCAGGGCTTGGCCCCGACGATACAGAAATGTCAGGAAGCGCGCGAAATGAAAAACATGGCGGTCAACAAAATGGCGGCGACATCAATACGGCTCTGAAACATTTAGATGAGATGCAGCAAGCCGATAAGAATGTCAGCAAAGCGATGATTTGTGATATTAGTAAACAAGTTGATACACAAATGAGTAAATTAGATCAGATGATTTGCCAAGCGGATCGCGCTCAGAATTCCTTGCAGCGACAAAATAAGCAGTTGAGGTCTTTCTTGCGATAG
Protein
MSLKAYEDACLRAELFGQPLPDKDEFIRKNKHLDIVEFEEVEIKTAENTAMLHDQMKEGTTGLTELNTILDSTQTKINRLKGVCGSITNFFRVKLNSKDNISYSSDPSYVGQTNFQRDYVPVSNRIDSVNAGLGPDDTEMSGSARNEKHGGQQNGGDINTALKHLDEMQQADKNVSKAMICDISKQVDTQMSKLDQMICQADRAQNSLQRQNKQLRSFLR
Summary
Similarity
Belongs to the BolA/IbaG family.
Uniprot
H9JTG1
A0A2H1WJ45
A0A2W1BH47
A0A3S2PD22
A0A212FMW4
A0A0L7L8C8
+ More
A0A194R1Q9 A0A194Q1W2 A0A2A4K4X6 A0A1L8DAH0 A0A1B0CRN9 A0A088A8H4 W8B6S6 A0A2A3EGJ4 A0A182SU19 A0A1Y1M6R4 A0A182M6M5 A0A182WR08 K7J950 A0A182V8C0 A0A182LE47 A0A182IIW2 A0A182S4P9 A0A182NZA4 A0A0J7KP32 Q7QCN2 A0A0L7REQ5 A0A182XUJ8 A0A1Q3G512 A0A2J7PGW2 A0A182Q094 A0A232F4L7 A0A182H2Y0 A0A310SDN8 E2AQ64 A0A182Y3Z8 A0A182JIL7 A0A195FPY4 A0A182K087 A0A084VRJ2 W5JKA7 Q17P73 A0A182TM18 A0A1S4EVW0 A0A026WX02 A0A1J1HJG5 E2B4H5 T1PJY9 A0A158NE90 A0A151J0F2 A0A195BX90 A0A034VIG1 A0A067QHS2 F4WJD8 E9IEL6 A0A2P8YGU1 A0A1A9WSX9 A0A1A9ZIC1 A0A1A9VRS0 A0A1I8NQA3 A0A195CAM7 A0A0A1WIH2 A0A1W4ULC3 A0A023EHC5 A0A1B0FE23 A0A336M7D0 A0A336LSB8 B4MTF4 B4I6Y5 A0A131Y0D3 V5IK54 A0A1B6DEK8 A0A0T6AU05 B7P3H7 A0A1B6BXY5 K7ITY9 K1QBK7 Q4RUU4
A0A194R1Q9 A0A194Q1W2 A0A2A4K4X6 A0A1L8DAH0 A0A1B0CRN9 A0A088A8H4 W8B6S6 A0A2A3EGJ4 A0A182SU19 A0A1Y1M6R4 A0A182M6M5 A0A182WR08 K7J950 A0A182V8C0 A0A182LE47 A0A182IIW2 A0A182S4P9 A0A182NZA4 A0A0J7KP32 Q7QCN2 A0A0L7REQ5 A0A182XUJ8 A0A1Q3G512 A0A2J7PGW2 A0A182Q094 A0A232F4L7 A0A182H2Y0 A0A310SDN8 E2AQ64 A0A182Y3Z8 A0A182JIL7 A0A195FPY4 A0A182K087 A0A084VRJ2 W5JKA7 Q17P73 A0A182TM18 A0A1S4EVW0 A0A026WX02 A0A1J1HJG5 E2B4H5 T1PJY9 A0A158NE90 A0A151J0F2 A0A195BX90 A0A034VIG1 A0A067QHS2 F4WJD8 E9IEL6 A0A2P8YGU1 A0A1A9WSX9 A0A1A9ZIC1 A0A1A9VRS0 A0A1I8NQA3 A0A195CAM7 A0A0A1WIH2 A0A1W4ULC3 A0A023EHC5 A0A1B0FE23 A0A336M7D0 A0A336LSB8 B4MTF4 B4I6Y5 A0A131Y0D3 V5IK54 A0A1B6DEK8 A0A0T6AU05 B7P3H7 A0A1B6BXY5 K7ITY9 K1QBK7 Q4RUU4
Pubmed
19121390
28756777
22118469
26227816
26354079
24495485
+ More
28004739 20075255 20966253 12364791 14747013 17210077 28648823 26483478 20798317 25244985 24438588 20920257 23761445 17510324 24508170 30249741 25315136 21347285 25348373 24845553 21719571 21282665 29403074 25830018 24945155 17994087 25765539 22992520 15496914
28004739 20075255 20966253 12364791 14747013 17210077 28648823 26483478 20798317 25244985 24438588 20920257 23761445 17510324 24508170 30249741 25315136 21347285 25348373 24845553 21719571 21282665 29403074 25830018 24945155 17994087 25765539 22992520 15496914
EMBL
BABH01003905
ODYU01009013
SOQ53100.1
KZ150097
PZC73591.1
RSAL01000093
+ More
RVE47925.1 AGBW02007651 OWR55030.1 JTDY01002361 KOB71584.1 KQ460878 KPJ11627.1 KQ459582 KPI98984.1 NWSH01000169 PCG78823.1 GFDF01010621 JAV03463.1 AJWK01025068 GAMC01013797 JAB92758.1 KZ288254 PBC30885.1 GEZM01040920 JAV80708.1 AXCM01000144 APCN01000245 LBMM01004860 KMQ92031.1 AAAB01008859 EAA07730.4 KQ414609 KOC69309.1 GFDL01000189 JAV34856.1 NEVH01025161 PNF15572.1 AXCN02001476 NNAY01001028 OXU25428.1 JXUM01024208 KQ560683 KXJ81286.1 KQ767320 OAD53413.1 GL441701 EFN64398.1 KQ981335 KYN42655.1 ATLV01015691 KE525027 KFB40586.1 ADMH02001265 ETN63320.1 CH477193 EAT48560.1 KK107078 QOIP01000011 EZA60368.1 RLU17226.1 CVRI01000002 CRK86614.1 GL445560 EFN89411.1 KA648410 AFP63039.1 ADTU01013175 KQ980636 KYN14932.1 KQ976401 KYM92578.1 GAKP01016693 GAKP01016691 JAC42259.1 KK853387 KDR07945.1 GL888182 EGI65716.1 GL762647 EFZ20989.1 PYGN01000604 PSN43480.1 KQ978023 KYM97924.1 GBXI01015811 JAC98480.1 GAPW01005344 JAC08254.1 CCAG010011618 UFQT01000659 SSX26224.1 UFQT01000150 SSX20906.1 CH963851 EDW75393.1 CH480823 EDW56083.1 GEFM01003077 JAP72719.1 GANP01000282 JAB84186.1 GEDC01013218 JAS24080.1 LJIG01022884 KRT78231.1 ABJB010128864 ABJB010574389 ABJB011056505 DS628877 EEC01149.1 GEDC01031180 JAS06118.1 JH818799 EKC28544.1 CAAE01014993 CAG07838.1
RVE47925.1 AGBW02007651 OWR55030.1 JTDY01002361 KOB71584.1 KQ460878 KPJ11627.1 KQ459582 KPI98984.1 NWSH01000169 PCG78823.1 GFDF01010621 JAV03463.1 AJWK01025068 GAMC01013797 JAB92758.1 KZ288254 PBC30885.1 GEZM01040920 JAV80708.1 AXCM01000144 APCN01000245 LBMM01004860 KMQ92031.1 AAAB01008859 EAA07730.4 KQ414609 KOC69309.1 GFDL01000189 JAV34856.1 NEVH01025161 PNF15572.1 AXCN02001476 NNAY01001028 OXU25428.1 JXUM01024208 KQ560683 KXJ81286.1 KQ767320 OAD53413.1 GL441701 EFN64398.1 KQ981335 KYN42655.1 ATLV01015691 KE525027 KFB40586.1 ADMH02001265 ETN63320.1 CH477193 EAT48560.1 KK107078 QOIP01000011 EZA60368.1 RLU17226.1 CVRI01000002 CRK86614.1 GL445560 EFN89411.1 KA648410 AFP63039.1 ADTU01013175 KQ980636 KYN14932.1 KQ976401 KYM92578.1 GAKP01016693 GAKP01016691 JAC42259.1 KK853387 KDR07945.1 GL888182 EGI65716.1 GL762647 EFZ20989.1 PYGN01000604 PSN43480.1 KQ978023 KYM97924.1 GBXI01015811 JAC98480.1 GAPW01005344 JAC08254.1 CCAG010011618 UFQT01000659 SSX26224.1 UFQT01000150 SSX20906.1 CH963851 EDW75393.1 CH480823 EDW56083.1 GEFM01003077 JAP72719.1 GANP01000282 JAB84186.1 GEDC01013218 JAS24080.1 LJIG01022884 KRT78231.1 ABJB010128864 ABJB010574389 ABJB011056505 DS628877 EEC01149.1 GEDC01031180 JAS06118.1 JH818799 EKC28544.1 CAAE01014993 CAG07838.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000037510
UP000053240
UP000053268
+ More
UP000218220 UP000092461 UP000005203 UP000242457 UP000075901 UP000075883 UP000075920 UP000002358 UP000075903 UP000075882 UP000075840 UP000075900 UP000075884 UP000036403 UP000007062 UP000053825 UP000076407 UP000235965 UP000075886 UP000215335 UP000069940 UP000249989 UP000000311 UP000076408 UP000075880 UP000078541 UP000075881 UP000030765 UP000000673 UP000008820 UP000075902 UP000053097 UP000279307 UP000183832 UP000008237 UP000095301 UP000005205 UP000078492 UP000078540 UP000027135 UP000007755 UP000245037 UP000091820 UP000092445 UP000078200 UP000095300 UP000078542 UP000192221 UP000092444 UP000007798 UP000001292 UP000001555 UP000005408 UP000007303
UP000218220 UP000092461 UP000005203 UP000242457 UP000075901 UP000075883 UP000075920 UP000002358 UP000075903 UP000075882 UP000075840 UP000075900 UP000075884 UP000036403 UP000007062 UP000053825 UP000076407 UP000235965 UP000075886 UP000215335 UP000069940 UP000249989 UP000000311 UP000076408 UP000075880 UP000078541 UP000075881 UP000030765 UP000000673 UP000008820 UP000075902 UP000053097 UP000279307 UP000183832 UP000008237 UP000095301 UP000005205 UP000078492 UP000078540 UP000027135 UP000007755 UP000245037 UP000091820 UP000092445 UP000078200 UP000095300 UP000078542 UP000192221 UP000092444 UP000007798 UP000001292 UP000001555 UP000005408 UP000007303
PRIDE
Interpro
ProteinModelPortal
H9JTG1
A0A2H1WJ45
A0A2W1BH47
A0A3S2PD22
A0A212FMW4
A0A0L7L8C8
+ More
A0A194R1Q9 A0A194Q1W2 A0A2A4K4X6 A0A1L8DAH0 A0A1B0CRN9 A0A088A8H4 W8B6S6 A0A2A3EGJ4 A0A182SU19 A0A1Y1M6R4 A0A182M6M5 A0A182WR08 K7J950 A0A182V8C0 A0A182LE47 A0A182IIW2 A0A182S4P9 A0A182NZA4 A0A0J7KP32 Q7QCN2 A0A0L7REQ5 A0A182XUJ8 A0A1Q3G512 A0A2J7PGW2 A0A182Q094 A0A232F4L7 A0A182H2Y0 A0A310SDN8 E2AQ64 A0A182Y3Z8 A0A182JIL7 A0A195FPY4 A0A182K087 A0A084VRJ2 W5JKA7 Q17P73 A0A182TM18 A0A1S4EVW0 A0A026WX02 A0A1J1HJG5 E2B4H5 T1PJY9 A0A158NE90 A0A151J0F2 A0A195BX90 A0A034VIG1 A0A067QHS2 F4WJD8 E9IEL6 A0A2P8YGU1 A0A1A9WSX9 A0A1A9ZIC1 A0A1A9VRS0 A0A1I8NQA3 A0A195CAM7 A0A0A1WIH2 A0A1W4ULC3 A0A023EHC5 A0A1B0FE23 A0A336M7D0 A0A336LSB8 B4MTF4 B4I6Y5 A0A131Y0D3 V5IK54 A0A1B6DEK8 A0A0T6AU05 B7P3H7 A0A1B6BXY5 K7ITY9 K1QBK7 Q4RUU4
A0A194R1Q9 A0A194Q1W2 A0A2A4K4X6 A0A1L8DAH0 A0A1B0CRN9 A0A088A8H4 W8B6S6 A0A2A3EGJ4 A0A182SU19 A0A1Y1M6R4 A0A182M6M5 A0A182WR08 K7J950 A0A182V8C0 A0A182LE47 A0A182IIW2 A0A182S4P9 A0A182NZA4 A0A0J7KP32 Q7QCN2 A0A0L7REQ5 A0A182XUJ8 A0A1Q3G512 A0A2J7PGW2 A0A182Q094 A0A232F4L7 A0A182H2Y0 A0A310SDN8 E2AQ64 A0A182Y3Z8 A0A182JIL7 A0A195FPY4 A0A182K087 A0A084VRJ2 W5JKA7 Q17P73 A0A182TM18 A0A1S4EVW0 A0A026WX02 A0A1J1HJG5 E2B4H5 T1PJY9 A0A158NE90 A0A151J0F2 A0A195BX90 A0A034VIG1 A0A067QHS2 F4WJD8 E9IEL6 A0A2P8YGU1 A0A1A9WSX9 A0A1A9ZIC1 A0A1A9VRS0 A0A1I8NQA3 A0A195CAM7 A0A0A1WIH2 A0A1W4ULC3 A0A023EHC5 A0A1B0FE23 A0A336M7D0 A0A336LSB8 B4MTF4 B4I6Y5 A0A131Y0D3 V5IK54 A0A1B6DEK8 A0A0T6AU05 B7P3H7 A0A1B6BXY5 K7ITY9 K1QBK7 Q4RUU4
Ontologies
GO
PANTHER
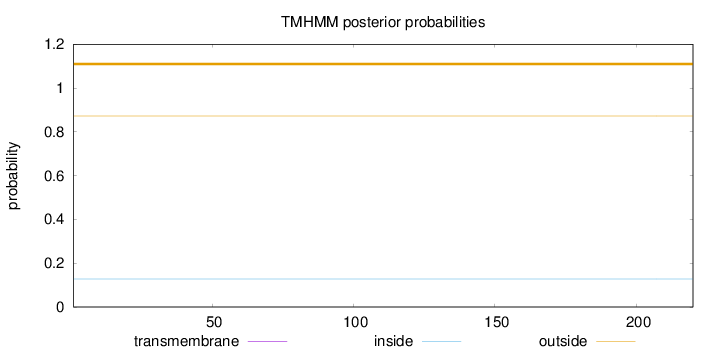
Topology
Length:
220
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00017
Exp number, first 60 AAs:
0
Total prob of N-in:
0.12807
outside
1 - 220
Population Genetic Test Statistics
Pi
278.273425
Theta
205.996212
Tajima's D
0.921881
CLR
0
CSRT
0.638618069096545
Interpretation
Uncertain
