Gene
KWMTBOMO09548 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013023
Annotation
PREDICTED:_bleomycin_hydrolase_isoform_X1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 3.053
Sequence
CDS
ATGTCGAGTTCAGTACCTCTAACATTAGATGAGCTATCAAAAATCCGCAAAGATTTTTATGCAGAACCAAAAAATGAATTGGCTCAGAATGTTTGTACTCGATATGATCCATTCGAAGTAGCCATCAACAGACGGAAGACAGATACTTCACTTCATGTTTTCAACCTTAAGATTGAGACTGAAGGGAAACCTGTCACCAATCAAGAGAGCTCCGGTCGATGCTGGCTGTTTGCAGCACTCAACGTGATGCGTTTGCCTTTCATGAAGAAATATGGAATTGAAGAATTTGAATTCTCACAAAGTTACCTGTTCTTTTGGGATAAGATTGAAAGATCACATTTCTGGTTGAACAACATCGCGGAGACCGCTAAAAAGGGGGAGAAACTTGACGGAAGAGTTGTGAACTTCTTGCTCAAGGATCCGGTCAACGACGGCGGCCAGTGGGACATGCTGGTGAACCTCGTGAACAAATACGGCTTAATGCCGAAGAAGTGCTTCCCGGAGTCGTACAGCTCGAGGCGGAGTGTGCGCATGAACGCGCTGCTGAGGACCAAGCTCCGTGAATTCGCCAAAGAATTGCGAGAAAAGGTAACGTCAGATGCATCCGACGACGAAATACAGAACACGATCAGTAAACAGATGATCGTCGTGTACAATATCGTAGCTATATGCTTGGGTATACCCCCGGAGAAGTTCACGTACGAGTATTACAATAAGGACAAGGCGTACCAGGTCATGGGTCCGCTGACGCCCCAGGAGTTCTACGCGCGGCACGTGAAGCCGCTGTACGACGTGGACGACAAGGTGTGCATAGTAAACGATCCACGAGAGAACAATCCGTATGGTCATTTATATACGCTACAGTATCTCGGTAATATGGTCGGAGGTCGAACGACTGTTTACAACAATCAGCCCATCGAAGTATTGATCAAGGCCGTTAAGGACAGCATCCAGGGAGGCGAAGCTGTGTGGTTCGGGTGTGAAGTGACGAAGAGGTTCGAACGGAAGAACGGCCTGGAGGATCTTGAAGCTCACGATTATCGTTTGGTCTTCAACACTGAGATCCAGATTGGGATGCCGAAGGAGGACCGACTGCTGTACGGAGACTCGTGCATGACGCACGCCATGGTGTTCACGGCGGTCGGCCTCGATGAGCAAGGCAACCCGCTGAAGTTCCGCGTCGAGAACTCTTCCAGCGACAAGGAGTACGATAAAGGCTACCTGCTACTCACCGAGCCTTGGTTCAGAGAGTTTGTGTTTGAGGTCGTTGTAGACAAGAAGTACGTGTCGAAAGAAGTTCTAGATGTGTTCAAACAAGAATTAGTGGAATTACCGGCCTGGGACCCCATGGGGACGCTGGCGTGCCCGCTCTGCGCGGACGACTAG
Protein
MSSSVPLTLDELSKIRKDFYAEPKNELAQNVCTRYDPFEVAINRRKTDTSLHVFNLKIETEGKPVTNQESSGRCWLFAALNVMRLPFMKKYGIEEFEFSQSYLFFWDKIERSHFWLNNIAETAKKGEKLDGRVVNFLLKDPVNDGGQWDMLVNLVNKYGLMPKKCFPESYSSRRSVRMNALLRTKLREFAKELREKVTSDASDDEIQNTISKQMIVVYNIVAICLGIPPEKFTYEYYNKDKAYQVMGPLTPQEFYARHVKPLYDVDDKVCIVNDPRENNPYGHLYTLQYLGNMVGGRTTVYNNQPIEVLIKAVKDSIQGGEAVWFGCEVTKRFERKNGLEDLEAHDYRLVFNTEIQIGMPKEDRLLYGDSCMTHAMVFTAVGLDEQGNPLKFRVENSSSDKEYDKGYLLLTEPWFREFVFEVVVDKKYVSKEVLDVFKQELVELPAWDPMGTLACPLCADD
Summary
Uniprot
H9JU11
A0A1E1W8V0
A0A2A4K569
A0A2W1BEP1
A0A194Q0Y9
A0A212FMR5
+ More
A0A194R6Y4 A0A3S2LJC1 A0A0K8TT30 A0A0L0C2J9 A0A2J7R829 K7J270 A0A1Y1KSS5 U4UXK3 A0A182JF13 A0A232EKI9 J3JXH8 A0A1W4X2M9 N6U1S1 A0A182VR47 A0A1A9XH54 A0A023ESH6 A0A182R4L5 A0A1A9VRS4 A0A182JQJ1 B0WKQ7 A0A182NSC2 A0A182QWH0 Q1HQS7 A0A084VS69 A0A1B0FE23 A0A182SNH3 W8C811 B4MSJ2 A0A1L8E2I6 A0A182PSG4 A0A182X9N6 W8BC54 A0A2C9GK43 A0A1I8JVI1 A0A182TU37 A0A182HKM0 Q7PKL9 A0A1L8ECS2 T1PC46 U5EUL6 A0A1L8EDP7 B4M2M8 A0A182MKX2 B3MVX9 A0A1Q3F8H2 A0A0Q9WNC7 B4R6I0 B4L4I0 A0A1Q3FD69 A0A0Q9WQ58 A0A0Q9WMP3 A0A1L8ED90 A8JV22 A0A0Q9WMP2 X2JEJ0 A0A0Q5T3K8 Q9W3F6 A0A034VER1 A0A182YGB0 A0A1I8Q0X7 W5J7F8 A0A034VI39 A0A0R1EBR1 B4IJV9 B4Q0B9 B3NVI3 A0A182F868 A0A2M4BN43 A0A0K8UZ36 A0A0R3P553 A0A2M3Z893 A0A2M3Z866 A0A0R3NZV9 A0A0R3NZR4 A0A1D2MPT2 A0A1W4VJ99 A0A0P4W7L8 A0A0K8VN73 A0A2M4BMQ2 A0A2M4BMQ3 Q29HJ1 A0A2M4AGA0 B4GXL0 A0A0A1XJK7 A0A2M4AG97 A0A182H8T9 T1DRF7 A0A2M4CHK1 A0A1A9WSY3 D2A4S5 A0A3B0KRM4 A0A3B0KL69 A0A3B0JWV4 A0A0M4EN33
A0A194R6Y4 A0A3S2LJC1 A0A0K8TT30 A0A0L0C2J9 A0A2J7R829 K7J270 A0A1Y1KSS5 U4UXK3 A0A182JF13 A0A232EKI9 J3JXH8 A0A1W4X2M9 N6U1S1 A0A182VR47 A0A1A9XH54 A0A023ESH6 A0A182R4L5 A0A1A9VRS4 A0A182JQJ1 B0WKQ7 A0A182NSC2 A0A182QWH0 Q1HQS7 A0A084VS69 A0A1B0FE23 A0A182SNH3 W8C811 B4MSJ2 A0A1L8E2I6 A0A182PSG4 A0A182X9N6 W8BC54 A0A2C9GK43 A0A1I8JVI1 A0A182TU37 A0A182HKM0 Q7PKL9 A0A1L8ECS2 T1PC46 U5EUL6 A0A1L8EDP7 B4M2M8 A0A182MKX2 B3MVX9 A0A1Q3F8H2 A0A0Q9WNC7 B4R6I0 B4L4I0 A0A1Q3FD69 A0A0Q9WQ58 A0A0Q9WMP3 A0A1L8ED90 A8JV22 A0A0Q9WMP2 X2JEJ0 A0A0Q5T3K8 Q9W3F6 A0A034VER1 A0A182YGB0 A0A1I8Q0X7 W5J7F8 A0A034VI39 A0A0R1EBR1 B4IJV9 B4Q0B9 B3NVI3 A0A182F868 A0A2M4BN43 A0A0K8UZ36 A0A0R3P553 A0A2M3Z893 A0A2M3Z866 A0A0R3NZV9 A0A0R3NZR4 A0A1D2MPT2 A0A1W4VJ99 A0A0P4W7L8 A0A0K8VN73 A0A2M4BMQ2 A0A2M4BMQ3 Q29HJ1 A0A2M4AGA0 B4GXL0 A0A0A1XJK7 A0A2M4AG97 A0A182H8T9 T1DRF7 A0A2M4CHK1 A0A1A9WSY3 D2A4S5 A0A3B0KRM4 A0A3B0KL69 A0A3B0JWV4 A0A0M4EN33
Pubmed
19121390
28756777
26354079
22118469
26369729
26108605
+ More
20075255 28004739 23537049 28648823 22516182 24945155 17204158 17510324 24438588 24495485 17994087 12364791 14747013 17210077 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 25244985 20920257 23761445 17550304 15632085 27289101 25830018 26483478 18362917 19820115
20075255 28004739 23537049 28648823 22516182 24945155 17204158 17510324 24438588 24495485 17994087 12364791 14747013 17210077 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 25244985 20920257 23761445 17550304 15632085 27289101 25830018 26483478 18362917 19820115
EMBL
BABH01003905
BABH01003906
BABH01003907
BABH01003908
GDQN01007658
JAT83396.1
+ More
NWSH01000169 PCG78822.1 KZ150097 PZC73592.1 KQ459582 KPI98983.1 AGBW02007651 OWR55031.1 KQ460878 KPJ11626.1 RSAL01000093 RVE47924.1 GDAI01000091 JAI17512.1 JRES01000984 KNC26457.1 NEVH01006723 PNF36988.1 AAZX01001579 GEZM01076803 JAV63638.1 KB632404 ERL95081.1 NNAY01003779 OXU18876.1 BT127947 AEE62909.1 APGK01052870 KB741216 ENN72502.1 GAPW01001380 JAC12218.1 DS231974 EDS29988.1 AXCN02000206 DQ440367 CH477482 ABF18400.1 EAT40213.1 ATLV01015825 KE525036 KFB40813.1 CCAG010011618 GAMC01007726 JAB98829.1 CH963851 EDW75081.2 GFDF01001157 JAV12927.1 GAMC01007725 JAB98830.1 APCN01000846 AAAB01008987 EAA43211.3 GFDG01002259 JAV16540.1 KA646347 AFP60976.1 GANO01002263 JAB57608.1 GFDG01002106 JAV16693.1 CH940651 EDW65932.2 AXCM01016291 CH902625 EDV35124.2 GFDL01011181 JAV23864.1 KRF82484.1 CM000366 EDX17410.1 CH933810 EDW07458.1 GFDL01009514 JAV25531.1 KRF82485.1 KRF94065.1 GFDG01002102 JAV16697.1 AE014298 BT120019 ABW09357.1 ACZ98471.1 KRF94066.1 AHN59489.1 CH954180 KQS29954.1 AY118343 AAF46371.2 AAM48372.1 GAKP01017171 JAC41781.1 ADMH02002026 ETN59896.1 GAKP01017170 JAC41782.1 CM000162 KRK06671.1 CH480850 EDW51315.1 EDX02256.2 EDV46375.1 GGFJ01005220 MBW54361.1 GDHF01020377 JAI31937.1 CH379064 KRT06515.1 GGFM01003959 MBW24710.1 GGFM01003944 MBW24695.1 KRT06516.1 KRT06514.1 LJIJ01000715 ODM95077.1 GDRN01076303 JAI62913.1 GDHF01012289 JAI40025.1 GGFJ01005216 MBW54357.1 GGFJ01005219 MBW54360.1 EAL31767.2 GGFK01006492 MBW39813.1 CH479196 EDW27487.1 GBXI01003589 JAD10703.1 GGFK01006493 MBW39814.1 JXUM01029657 JXUM01029658 KQ560860 KXJ80601.1 GAMD01000645 JAB00946.1 GGFL01000626 MBW64804.1 KQ971344 EFA05186.1 OUUW01000011 SPP86568.1 SPP86566.1 SPP86567.1 CP012528 ALC49308.1
NWSH01000169 PCG78822.1 KZ150097 PZC73592.1 KQ459582 KPI98983.1 AGBW02007651 OWR55031.1 KQ460878 KPJ11626.1 RSAL01000093 RVE47924.1 GDAI01000091 JAI17512.1 JRES01000984 KNC26457.1 NEVH01006723 PNF36988.1 AAZX01001579 GEZM01076803 JAV63638.1 KB632404 ERL95081.1 NNAY01003779 OXU18876.1 BT127947 AEE62909.1 APGK01052870 KB741216 ENN72502.1 GAPW01001380 JAC12218.1 DS231974 EDS29988.1 AXCN02000206 DQ440367 CH477482 ABF18400.1 EAT40213.1 ATLV01015825 KE525036 KFB40813.1 CCAG010011618 GAMC01007726 JAB98829.1 CH963851 EDW75081.2 GFDF01001157 JAV12927.1 GAMC01007725 JAB98830.1 APCN01000846 AAAB01008987 EAA43211.3 GFDG01002259 JAV16540.1 KA646347 AFP60976.1 GANO01002263 JAB57608.1 GFDG01002106 JAV16693.1 CH940651 EDW65932.2 AXCM01016291 CH902625 EDV35124.2 GFDL01011181 JAV23864.1 KRF82484.1 CM000366 EDX17410.1 CH933810 EDW07458.1 GFDL01009514 JAV25531.1 KRF82485.1 KRF94065.1 GFDG01002102 JAV16697.1 AE014298 BT120019 ABW09357.1 ACZ98471.1 KRF94066.1 AHN59489.1 CH954180 KQS29954.1 AY118343 AAF46371.2 AAM48372.1 GAKP01017171 JAC41781.1 ADMH02002026 ETN59896.1 GAKP01017170 JAC41782.1 CM000162 KRK06671.1 CH480850 EDW51315.1 EDX02256.2 EDV46375.1 GGFJ01005220 MBW54361.1 GDHF01020377 JAI31937.1 CH379064 KRT06515.1 GGFM01003959 MBW24710.1 GGFM01003944 MBW24695.1 KRT06516.1 KRT06514.1 LJIJ01000715 ODM95077.1 GDRN01076303 JAI62913.1 GDHF01012289 JAI40025.1 GGFJ01005216 MBW54357.1 GGFJ01005219 MBW54360.1 EAL31767.2 GGFK01006492 MBW39813.1 CH479196 EDW27487.1 GBXI01003589 JAD10703.1 GGFK01006493 MBW39814.1 JXUM01029657 JXUM01029658 KQ560860 KXJ80601.1 GAMD01000645 JAB00946.1 GGFL01000626 MBW64804.1 KQ971344 EFA05186.1 OUUW01000011 SPP86568.1 SPP86566.1 SPP86567.1 CP012528 ALC49308.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000283053
+ More
UP000037069 UP000235965 UP000002358 UP000030742 UP000075880 UP000215335 UP000192223 UP000019118 UP000075920 UP000092443 UP000075900 UP000078200 UP000075881 UP000002320 UP000075884 UP000075886 UP000008820 UP000030765 UP000092444 UP000075901 UP000007798 UP000075885 UP000076407 UP000075840 UP000075903 UP000075902 UP000007062 UP000095301 UP000008792 UP000075883 UP000007801 UP000000304 UP000009192 UP000000803 UP000008711 UP000076408 UP000095300 UP000000673 UP000002282 UP000001292 UP000069272 UP000001819 UP000094527 UP000192221 UP000008744 UP000069940 UP000249989 UP000091820 UP000007266 UP000268350 UP000092553
UP000037069 UP000235965 UP000002358 UP000030742 UP000075880 UP000215335 UP000192223 UP000019118 UP000075920 UP000092443 UP000075900 UP000078200 UP000075881 UP000002320 UP000075884 UP000075886 UP000008820 UP000030765 UP000092444 UP000075901 UP000007798 UP000075885 UP000076407 UP000075840 UP000075903 UP000075902 UP000007062 UP000095301 UP000008792 UP000075883 UP000007801 UP000000304 UP000009192 UP000000803 UP000008711 UP000076408 UP000095300 UP000000673 UP000002282 UP000001292 UP000069272 UP000001819 UP000094527 UP000192221 UP000008744 UP000069940 UP000249989 UP000091820 UP000007266 UP000268350 UP000092553
Interpro
SUPFAM
SSF54001
SSF54001
Gene 3D
ProteinModelPortal
H9JU11
A0A1E1W8V0
A0A2A4K569
A0A2W1BEP1
A0A194Q0Y9
A0A212FMR5
+ More
A0A194R6Y4 A0A3S2LJC1 A0A0K8TT30 A0A0L0C2J9 A0A2J7R829 K7J270 A0A1Y1KSS5 U4UXK3 A0A182JF13 A0A232EKI9 J3JXH8 A0A1W4X2M9 N6U1S1 A0A182VR47 A0A1A9XH54 A0A023ESH6 A0A182R4L5 A0A1A9VRS4 A0A182JQJ1 B0WKQ7 A0A182NSC2 A0A182QWH0 Q1HQS7 A0A084VS69 A0A1B0FE23 A0A182SNH3 W8C811 B4MSJ2 A0A1L8E2I6 A0A182PSG4 A0A182X9N6 W8BC54 A0A2C9GK43 A0A1I8JVI1 A0A182TU37 A0A182HKM0 Q7PKL9 A0A1L8ECS2 T1PC46 U5EUL6 A0A1L8EDP7 B4M2M8 A0A182MKX2 B3MVX9 A0A1Q3F8H2 A0A0Q9WNC7 B4R6I0 B4L4I0 A0A1Q3FD69 A0A0Q9WQ58 A0A0Q9WMP3 A0A1L8ED90 A8JV22 A0A0Q9WMP2 X2JEJ0 A0A0Q5T3K8 Q9W3F6 A0A034VER1 A0A182YGB0 A0A1I8Q0X7 W5J7F8 A0A034VI39 A0A0R1EBR1 B4IJV9 B4Q0B9 B3NVI3 A0A182F868 A0A2M4BN43 A0A0K8UZ36 A0A0R3P553 A0A2M3Z893 A0A2M3Z866 A0A0R3NZV9 A0A0R3NZR4 A0A1D2MPT2 A0A1W4VJ99 A0A0P4W7L8 A0A0K8VN73 A0A2M4BMQ2 A0A2M4BMQ3 Q29HJ1 A0A2M4AGA0 B4GXL0 A0A0A1XJK7 A0A2M4AG97 A0A182H8T9 T1DRF7 A0A2M4CHK1 A0A1A9WSY3 D2A4S5 A0A3B0KRM4 A0A3B0KL69 A0A3B0JWV4 A0A0M4EN33
A0A194R6Y4 A0A3S2LJC1 A0A0K8TT30 A0A0L0C2J9 A0A2J7R829 K7J270 A0A1Y1KSS5 U4UXK3 A0A182JF13 A0A232EKI9 J3JXH8 A0A1W4X2M9 N6U1S1 A0A182VR47 A0A1A9XH54 A0A023ESH6 A0A182R4L5 A0A1A9VRS4 A0A182JQJ1 B0WKQ7 A0A182NSC2 A0A182QWH0 Q1HQS7 A0A084VS69 A0A1B0FE23 A0A182SNH3 W8C811 B4MSJ2 A0A1L8E2I6 A0A182PSG4 A0A182X9N6 W8BC54 A0A2C9GK43 A0A1I8JVI1 A0A182TU37 A0A182HKM0 Q7PKL9 A0A1L8ECS2 T1PC46 U5EUL6 A0A1L8EDP7 B4M2M8 A0A182MKX2 B3MVX9 A0A1Q3F8H2 A0A0Q9WNC7 B4R6I0 B4L4I0 A0A1Q3FD69 A0A0Q9WQ58 A0A0Q9WMP3 A0A1L8ED90 A8JV22 A0A0Q9WMP2 X2JEJ0 A0A0Q5T3K8 Q9W3F6 A0A034VER1 A0A182YGB0 A0A1I8Q0X7 W5J7F8 A0A034VI39 A0A0R1EBR1 B4IJV9 B4Q0B9 B3NVI3 A0A182F868 A0A2M4BN43 A0A0K8UZ36 A0A0R3P553 A0A2M3Z893 A0A2M3Z866 A0A0R3NZV9 A0A0R3NZR4 A0A1D2MPT2 A0A1W4VJ99 A0A0P4W7L8 A0A0K8VN73 A0A2M4BMQ2 A0A2M4BMQ3 Q29HJ1 A0A2M4AGA0 B4GXL0 A0A0A1XJK7 A0A2M4AG97 A0A182H8T9 T1DRF7 A0A2M4CHK1 A0A1A9WSY3 D2A4S5 A0A3B0KRM4 A0A3B0KL69 A0A3B0JWV4 A0A0M4EN33
PDB
1CB5
E-value=6.05041e-145,
Score=1319
Ontologies
GO
PANTHER
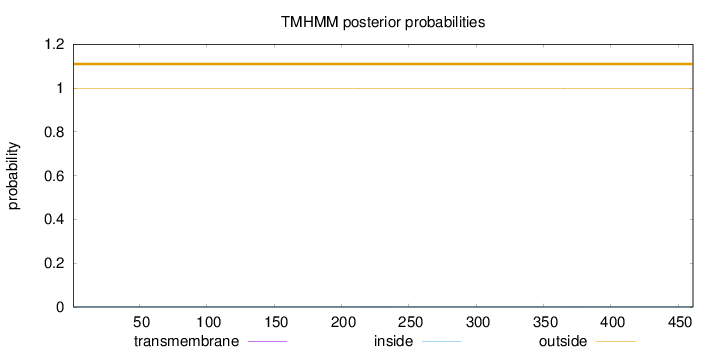
Topology
Length:
461
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03864
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00167
outside
1 - 461
Population Genetic Test Statistics
Pi
213.829836
Theta
153.701111
Tajima's D
1.562987
CLR
0.176797
CSRT
0.800409979501025
Interpretation
Uncertain
