Gene
KWMTBOMO09545
Pre Gene Modal
BGIBMGA013024
Annotation
PREDICTED:_nose_resistant_to_fluoxetine_protein_6-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.744
Sequence
CDS
ATGTCAGCGAGTGAATTTACAGATGAAGAGTACTATCACCTGCCGGATCTGTACCATTTGGACGACTACACGAAATGTCTGTCGCAGAAACAGGGCCTGTATTGCCTGGGAACTTTCGAACTGTCACCAGCTGAAGAACAGAGTCCAGTTTTCGATCTTATCAAGGTGTATTCAGAAAACCCTCATCACTTCAACCGCACGATCATACAGCGCGGCTGGTGCGTCAGCAGTCGCAGCCTAAGAACCGAAGCCGAGCCGGTGCGCAGATTCACGAAATGCGGTCAAGATTGGGCCCGCTCCCGGGGGATGAACGCTCGCCTGCTAAAACTCGAATATTGCCGAACTCACGATGAAGCAAAAACTCCGAACGCTTCGGACTTGCCAGAAAGAGTGTTCCTCTGTGCAATTATCTGTATTCTAATCTTCAACGTCATCGGAACGGCCTACGATTTATGGAAAGGAAACGAAGAAAACAAAAACAAAATTCTAATGGCGTGGTCTATGCCCAGCAACTGGTCGCGCTTGACGTCATCCTTGTCGAATGAAGATCCTCGACTTTCATGCTTAGCGCCGTTTCAGGGAGCAAGAGTCATCGTAATGACCTTGGTGCTGTGGGGTCATGTGTTTTTCTCACACGCGTTGGTGTATCTCCAGAACCCACGAGATTACGAAAAATCTCTCCACGGCGTTATTGGCGCACTGGTTTATAATGGGACATCCATAGTGCAGATGTTCGTTATTTTTTCAAACTTCCTCTGCGCATACAATCTACTATTGCCGTCAAATAAAAAACCGCTCACGCTAAGCATGTTCCCGAAGCAAATCGTCAAGAGATATAGCAGAATAACTCCTATGAACTTGCTCCTGGTGGGCTACGGAGCCACGTGGTGGGTTCATAGCCGCGACGGTGCATTATGGCCGGTCCTAGTTGGAAACGAGAAGATTTTTTGCGAACAAAAGTTCTGGTCTCATGCGCTGTACATCAATAACTTTTACATGCCTGAAAACGTTTGTTTCATACAGACATGGTTCTTAGCGGTTGACATGCAGCTCCATATCCTAGCTCTCATTCTGACTTTGTGTCTGGCTAAGCACCGACGCCATGCCCTACGCATCCTCGGAAGCCTGTTCGTACTATCCTGTGCTGGTCTAAGCTTAATGGTGTACTTAAACAACTACTCGACGTTATTACAAATCGCTGAACCTGATAATGCACGCACGATGCTCCGTAATAAGCCGTCGTTTTACGAAATGTACGTGAATCCATTTAACTCGCTACCAGCTTGCCTGATGGGTTTGTTTCTGGCTCATCTACTTTACGAATTACAAGAAAACGGATATAAATTTCACGAATGCAAATGGCTGAAGTACTCTCACGCTTTAATGATACCGCTGTACATATTATGGCTGTTTTGCGGGATTTATTTCAGAGAACTAAAATCTAATATAGACACAGCTCTATTCATTGGATTGGATCGACTGCTATTCTGTTCAATATTCGCAGTAAAACTAATGGGATTTACGACAATTCAATATTTCCCAACCTTCATACCGGAACAGAGGATTGCTTTGCAGCACAAATAG
Protein
MSASEFTDEEYYHLPDLYHLDDYTKCLSQKQGLYCLGTFELSPAEEQSPVFDLIKVYSENPHHFNRTIIQRGWCVSSRSLRTEAEPVRRFTKCGQDWARSRGMNARLLKLEYCRTHDEAKTPNASDLPERVFLCAIICILIFNVIGTAYDLWKGNEENKNKILMAWSMPSNWSRLTSSLSNEDPRLSCLAPFQGARVIVMTLVLWGHVFFSHALVYLQNPRDYEKSLHGVIGALVYNGTSIVQMFVIFSNFLCAYNLLLPSNKKPLTLSMFPKQIVKRYSRITPMNLLLVGYGATWWVHSRDGALWPVLVGNEKIFCEQKFWSHALYINNFYMPENVCFIQTWFLAVDMQLHILALILTLCLAKHRRHALRILGSLFVLSCAGLSLMVYLNNYSTLLQIAEPDNARTMLRNKPSFYEMYVNPFNSLPACLMGLFLAHLLYELQENGYKFHECKWLKYSHALMIPLYILWLFCGIYFRELKSNIDTALFIGLDRLLFCSIFAVKLMGFTTIQYFPTFIPEQRIALQHK
Summary
Catalytic Activity
S-ubiquitinyl-[E2 ubiquitin-conjugating enzyme]-L-cysteine + [acceptor protein]-L-lysine = [E2 ubiquitin-conjugating enzyme]-L-cysteine + N(6)-ubiquitinyl-[acceptor protein]-L-lysine.
Uniprot
A0A2A4K959
H9JU90
A0A2W1BKH9
A0A212FM91
A0A2A4K429
A0A2W1BIR4
+ More
A0A2H1WLB9 A0A2H1VFZ7 A0A0N0PCP5 A0A2A4K862 A0A194Q086 A0A2W1BRV2 A0A0N1IH78 A0A2A4K7P3 A0A194Q6D5 A0A212FMC6 A0A3S2P8D1 A0A212ETZ7 A0A437AY35 A0A2H1VKS3 A0A0N1IBJ6 A0A194QFD0 A0A2H1WAT5 A0A0N1I4J5 A0A212FM81 A0A2W1BKH3 A0A194Q1V4 A0A0N0P9S1 A0A2H1WAR4 A0A2A4JGF7 A0A212FGA7 A0A1Y1M2Q0 A0A2W1BK32 A0A2A4JGW2 A0A2W1BK62 A0A0N1ID86 A0A0N0PAP1 A0A2H1WCA0 A0A2W1BKB8 A0A194QKZ1 A0A1Y1KVX9 B0WNX6 A0A2H1WCF8 M4M6W5 A0A0L7KR23 A0A1W4VFG7 A0A182VP89 A0A1S4GUU3 A0A182KQ82 Q7QIT8 A0A1S4FD62 A0A182WWU1 A0A212EGP9 A0A1W4V1V5 A0A194QF68 A0A1S4FTS9 A0A182I2D7 A0A1Y1K4C2 Q16PV8 A0A182R1B6 B3MGQ2 A0A2J7Q6G4 A0A2H1WAT4 A0A182YEG1 Q176L6 A0A182NJC1 A0A194QVJ9 A0A1A9WWJ5 A0A182WLD5 A0A182TKI8 A0A084WEE0 A0A0A1X2L2 A0A182QFB6 Q7PTG8 A0A182PUZ7 A0A0J9R828 A0A182R4J9 A0A182MV59 A1Z6Q3 A0A1J1I7L1 A0A084WED7
A0A2H1WLB9 A0A2H1VFZ7 A0A0N0PCP5 A0A2A4K862 A0A194Q086 A0A2W1BRV2 A0A0N1IH78 A0A2A4K7P3 A0A194Q6D5 A0A212FMC6 A0A3S2P8D1 A0A212ETZ7 A0A437AY35 A0A2H1VKS3 A0A0N1IBJ6 A0A194QFD0 A0A2H1WAT5 A0A0N1I4J5 A0A212FM81 A0A2W1BKH3 A0A194Q1V4 A0A0N0P9S1 A0A2H1WAR4 A0A2A4JGF7 A0A212FGA7 A0A1Y1M2Q0 A0A2W1BK32 A0A2A4JGW2 A0A2W1BK62 A0A0N1ID86 A0A0N0PAP1 A0A2H1WCA0 A0A2W1BKB8 A0A194QKZ1 A0A1Y1KVX9 B0WNX6 A0A2H1WCF8 M4M6W5 A0A0L7KR23 A0A1W4VFG7 A0A182VP89 A0A1S4GUU3 A0A182KQ82 Q7QIT8 A0A1S4FD62 A0A182WWU1 A0A212EGP9 A0A1W4V1V5 A0A194QF68 A0A1S4FTS9 A0A182I2D7 A0A1Y1K4C2 Q16PV8 A0A182R1B6 B3MGQ2 A0A2J7Q6G4 A0A2H1WAT4 A0A182YEG1 Q176L6 A0A182NJC1 A0A194QVJ9 A0A1A9WWJ5 A0A182WLD5 A0A182TKI8 A0A084WEE0 A0A0A1X2L2 A0A182QFB6 Q7PTG8 A0A182PUZ7 A0A0J9R828 A0A182R4J9 A0A182MV59 A1Z6Q3 A0A1J1I7L1 A0A084WED7
Pubmed
EMBL
NWSH01000045
PCG80260.1
BABH01004270
BABH01004271
KZ149997
PZC75398.1
+ More
AGBW02007654 OWR54851.1 NWSH01000169 PCG78819.1 KZ150097 PZC73595.1 ODYU01009435 SOQ53859.1 ODYU01002360 SOQ39759.1 KQ460635 KPJ13420.1 PCG80259.1 KQ459582 KPI98971.1 PZC75396.1 KPJ13421.1 PCG80257.1 KPI98970.1 OWR54850.1 RSAL01000205 RVE44373.1 AGBW02012502 OWR44975.1 RSAL01000280 RVE43059.1 ODYU01003108 SOQ41449.1 KQ461184 KPJ07525.1 KQ459053 KPJ04119.1 ODYU01007420 SOQ50170.1 KQ459449 KPJ00879.1 OWR54852.1 KZ150085 PZC73777.1 KPI98974.1 KPJ00884.1 SOQ50171.1 NWSH01001667 PCG70472.1 AGBW02008687 OWR52781.1 GEZM01042633 JAV79751.1 PZC75399.1 NWSH01001633 PCG70632.1 KZ150068 PZC74095.1 KPJ07535.1 KPJ07529.1 ODYU01007576 SOQ50462.1 PZC74094.1 KPJ04116.1 GEZM01074205 GEZM01074204 JAV64641.1 DS232017 EDS31995.1 SOQ50174.1 JX846987 AGG55004.1 JTDY01006970 KOB65560.1 AAAB01008807 EAA04081.4 AGBW02015045 OWR40663.1 KPJ04127.1 APCN01000146 GEZM01096350 JAV54960.1 CH477771 EAT36390.1 AXCN02000448 AXCN02000449 CH902619 EDV36810.2 NEVH01017478 PNF24171.1 SOQ50173.1 CH477386 EAT42065.1 KQ461073 KPJ09497.1 ATLV01023205 KE525341 KFB48584.1 GBXI01009399 JAD04893.1 EAA03947.5 CM002911 KMY91834.1 AXCM01001275 AE013599 AAF57404.4 CVRI01000043 CRK96295.1 ATLV01023204 KFB48581.1
AGBW02007654 OWR54851.1 NWSH01000169 PCG78819.1 KZ150097 PZC73595.1 ODYU01009435 SOQ53859.1 ODYU01002360 SOQ39759.1 KQ460635 KPJ13420.1 PCG80259.1 KQ459582 KPI98971.1 PZC75396.1 KPJ13421.1 PCG80257.1 KPI98970.1 OWR54850.1 RSAL01000205 RVE44373.1 AGBW02012502 OWR44975.1 RSAL01000280 RVE43059.1 ODYU01003108 SOQ41449.1 KQ461184 KPJ07525.1 KQ459053 KPJ04119.1 ODYU01007420 SOQ50170.1 KQ459449 KPJ00879.1 OWR54852.1 KZ150085 PZC73777.1 KPI98974.1 KPJ00884.1 SOQ50171.1 NWSH01001667 PCG70472.1 AGBW02008687 OWR52781.1 GEZM01042633 JAV79751.1 PZC75399.1 NWSH01001633 PCG70632.1 KZ150068 PZC74095.1 KPJ07535.1 KPJ07529.1 ODYU01007576 SOQ50462.1 PZC74094.1 KPJ04116.1 GEZM01074205 GEZM01074204 JAV64641.1 DS232017 EDS31995.1 SOQ50174.1 JX846987 AGG55004.1 JTDY01006970 KOB65560.1 AAAB01008807 EAA04081.4 AGBW02015045 OWR40663.1 KPJ04127.1 APCN01000146 GEZM01096350 JAV54960.1 CH477771 EAT36390.1 AXCN02000448 AXCN02000449 CH902619 EDV36810.2 NEVH01017478 PNF24171.1 SOQ50173.1 CH477386 EAT42065.1 KQ461073 KPJ09497.1 ATLV01023205 KE525341 KFB48584.1 GBXI01009399 JAD04893.1 EAA03947.5 CM002911 KMY91834.1 AXCM01001275 AE013599 AAF57404.4 CVRI01000043 CRK96295.1 ATLV01023204 KFB48581.1
Proteomes
UP000218220
UP000005204
UP000007151
UP000053240
UP000053268
UP000283053
+ More
UP000002320 UP000037510 UP000192221 UP000075903 UP000075882 UP000007062 UP000076407 UP000075840 UP000008820 UP000075886 UP000007801 UP000235965 UP000076408 UP000075884 UP000091820 UP000075920 UP000075902 UP000030765 UP000075885 UP000075900 UP000075883 UP000000803 UP000183832
UP000002320 UP000037510 UP000192221 UP000075903 UP000075882 UP000007062 UP000076407 UP000075840 UP000008820 UP000075886 UP000007801 UP000235965 UP000076408 UP000075884 UP000091820 UP000075920 UP000075902 UP000030765 UP000075885 UP000075900 UP000075883 UP000000803 UP000183832
Pfam
Interpro
IPR002656
Acyl_transf_3
+ More
IPR009003 Peptidase_S1_PA
IPR001254 Trypsin_dom
IPR004119 EcKinase-like
IPR011009 Kinase-like_dom_sf
IPR015897 CHK_kinase-like
IPR007577 GlycoTrfase_DXD_sugar-bd_CS
IPR000569 HECT_dom
IPR035983 Hect_E3_ubiquitin_ligase
IPR035892 C2_domain_sf
IPR036020 WW_dom_sf
IPR001202 WW_dom
IPR000008 C2_dom
IPR023803 Ribosomal_S16_dom_sf
IPR008979 Galactose-bd-like_sf
IPR005201 Glyco_hydro_85
IPR000307 Ribosomal_S16
IPR009003 Peptidase_S1_PA
IPR001254 Trypsin_dom
IPR004119 EcKinase-like
IPR011009 Kinase-like_dom_sf
IPR015897 CHK_kinase-like
IPR007577 GlycoTrfase_DXD_sugar-bd_CS
IPR000569 HECT_dom
IPR035983 Hect_E3_ubiquitin_ligase
IPR035892 C2_domain_sf
IPR036020 WW_dom_sf
IPR001202 WW_dom
IPR000008 C2_dom
IPR023803 Ribosomal_S16_dom_sf
IPR008979 Galactose-bd-like_sf
IPR005201 Glyco_hydro_85
IPR000307 Ribosomal_S16
SUPFAM
Gene 3D
ProteinModelPortal
A0A2A4K959
H9JU90
A0A2W1BKH9
A0A212FM91
A0A2A4K429
A0A2W1BIR4
+ More
A0A2H1WLB9 A0A2H1VFZ7 A0A0N0PCP5 A0A2A4K862 A0A194Q086 A0A2W1BRV2 A0A0N1IH78 A0A2A4K7P3 A0A194Q6D5 A0A212FMC6 A0A3S2P8D1 A0A212ETZ7 A0A437AY35 A0A2H1VKS3 A0A0N1IBJ6 A0A194QFD0 A0A2H1WAT5 A0A0N1I4J5 A0A212FM81 A0A2W1BKH3 A0A194Q1V4 A0A0N0P9S1 A0A2H1WAR4 A0A2A4JGF7 A0A212FGA7 A0A1Y1M2Q0 A0A2W1BK32 A0A2A4JGW2 A0A2W1BK62 A0A0N1ID86 A0A0N0PAP1 A0A2H1WCA0 A0A2W1BKB8 A0A194QKZ1 A0A1Y1KVX9 B0WNX6 A0A2H1WCF8 M4M6W5 A0A0L7KR23 A0A1W4VFG7 A0A182VP89 A0A1S4GUU3 A0A182KQ82 Q7QIT8 A0A1S4FD62 A0A182WWU1 A0A212EGP9 A0A1W4V1V5 A0A194QF68 A0A1S4FTS9 A0A182I2D7 A0A1Y1K4C2 Q16PV8 A0A182R1B6 B3MGQ2 A0A2J7Q6G4 A0A2H1WAT4 A0A182YEG1 Q176L6 A0A182NJC1 A0A194QVJ9 A0A1A9WWJ5 A0A182WLD5 A0A182TKI8 A0A084WEE0 A0A0A1X2L2 A0A182QFB6 Q7PTG8 A0A182PUZ7 A0A0J9R828 A0A182R4J9 A0A182MV59 A1Z6Q3 A0A1J1I7L1 A0A084WED7
A0A2H1WLB9 A0A2H1VFZ7 A0A0N0PCP5 A0A2A4K862 A0A194Q086 A0A2W1BRV2 A0A0N1IH78 A0A2A4K7P3 A0A194Q6D5 A0A212FMC6 A0A3S2P8D1 A0A212ETZ7 A0A437AY35 A0A2H1VKS3 A0A0N1IBJ6 A0A194QFD0 A0A2H1WAT5 A0A0N1I4J5 A0A212FM81 A0A2W1BKH3 A0A194Q1V4 A0A0N0P9S1 A0A2H1WAR4 A0A2A4JGF7 A0A212FGA7 A0A1Y1M2Q0 A0A2W1BK32 A0A2A4JGW2 A0A2W1BK62 A0A0N1ID86 A0A0N0PAP1 A0A2H1WCA0 A0A2W1BKB8 A0A194QKZ1 A0A1Y1KVX9 B0WNX6 A0A2H1WCF8 M4M6W5 A0A0L7KR23 A0A1W4VFG7 A0A182VP89 A0A1S4GUU3 A0A182KQ82 Q7QIT8 A0A1S4FD62 A0A182WWU1 A0A212EGP9 A0A1W4V1V5 A0A194QF68 A0A1S4FTS9 A0A182I2D7 A0A1Y1K4C2 Q16PV8 A0A182R1B6 B3MGQ2 A0A2J7Q6G4 A0A2H1WAT4 A0A182YEG1 Q176L6 A0A182NJC1 A0A194QVJ9 A0A1A9WWJ5 A0A182WLD5 A0A182TKI8 A0A084WEE0 A0A0A1X2L2 A0A182QFB6 Q7PTG8 A0A182PUZ7 A0A0J9R828 A0A182R4J9 A0A182MV59 A1Z6Q3 A0A1J1I7L1 A0A084WED7
Ontologies
GO
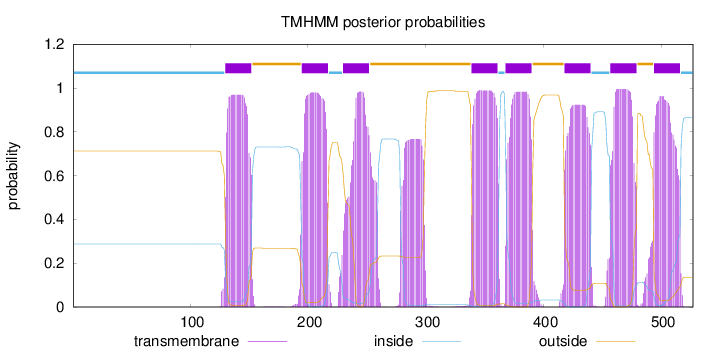
Topology
Length:
527
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
186.91167
Exp number, first 60 AAs:
0.0002
Total prob of N-in:
0.28802
inside
1 - 129
TMhelix
130 - 152
outside
153 - 194
TMhelix
195 - 217
inside
218 - 229
TMhelix
230 - 252
outside
253 - 338
TMhelix
339 - 361
inside
362 - 367
TMhelix
368 - 390
outside
391 - 417
TMhelix
418 - 440
inside
441 - 456
TMhelix
457 - 479
outside
480 - 493
TMhelix
494 - 516
inside
517 - 527
Population Genetic Test Statistics
Pi
200.815303
Theta
169.014608
Tajima's D
0.310457
CLR
0.66217
CSRT
0.458077096145193
Interpretation
Uncertain
