Gene
KWMTBOMO09544
Pre Gene Modal
BGIBMGA012819
Annotation
PREDICTED:_uncharacterized_protein_LOC105842067_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.966
Sequence
CDS
ATGTTGGTTAGTCTGTGTCTGTTGGCAGCACTCATCGAAGTCAAAGCGGCTGATTTTAATGGGGAAGATTACTGGCTTCTTCCTCGTCTCTTCGATTTGGACAACTACGATGAGTGTCTTTCGCATCCCGACGGCTTGTACTGCCTCGGCAGCTTCCATCTCACACCTGCTGAACAACCGAACCGTGTTTACGAATTGATGACTAACTATTCAAAGGAATTGTATAGGTTCAATCGCACTGTTATCCGACGTGGATGGTGTGTTAGTGCACGTGAATCGAATGTAACAGCGGAGCCGATGAATAGGTTTGAGCACTGCGCCGCGGAGTGGGCCCGCTCTCGTCGCATGCGCGCCCGTCTACTCAGACTGGAGTACTGCAGGAGCCATAGCGACGTACACGCCACACGGATCCCTGACAAGTCCGAACTAACCTTTCTCATTGCGATAGCTGTGATAATATTATTGAACGCAGCCAGTACCGCGTATGATTACGCCACTCAGAATAATGAGAAAAAGATTGCTCTCCTAACAACCTGGTCTGTTGTCGCCAACTGGAGGCAACTAGTTGAACCTTATAACTCACATCGGAAATCTCTACAAGCAATCCAAGGAATAAAAGTTATAGTTTCTGCGAGTATCTGTATTGTGCATAGTGGAATGTTGTTATATACATTGTTCTTGTATGAACCGATAGAAATGGAGCAGGTAATAGCGAATCCATTAACAATGGCGGTCCAGAACAGTACGACGGTCGTGCACATGTTCATTGTACTGTCAAGTTTCCTTACTGTTCACAATCTGTTAAAGTCAAGCAAACCGTTTGGTTTCGGTACATTCTTAAGGTTGCTCGCGAAAAGAATAATTAGATTAAGCCCTGTGTGTCTATTGGTGATCGGTTTCGCCGGGACCTGGTGGCCGCGGGTCAGTGACGGACCTCTGTGGCCGGCTGCGGTCGCGGAGCAGAGTAAGATCTGCCGCGAGAAGTTCTGGGCGCACGCGTTCTTTATCCATAACCTGGTGGAACCGGACAAGTTCTGTTTGATACCGACCTGGTATATAGCGGTCGACATGCAGATGTACATACTGGCAGTTATCTTAACAATGATCCTGGCCAAGTTCAGTAAGAAGGCTGTGCCGATTCTCGTGATTCTATTCATAGGAGCCACAGCGTTGAATGCGGGGTTGGCGTACATATACGAATGGAAGTCATTGTTGTATTTGGCAATAGTCGCAGAAAATATCAACTCTATATTCCGTGATATTCCATCATTGAGCCACTTTTACATGTCGCCTTGGGGCAGTCTTCCAGCGTGTCTCGTGGGACTCATCATAGGACATTTGGTCCATTTTACTAAGGGAGAAACATTTGGAAACATATACAAGCGACGGCTCCTCAGCTTATATTACGTTCTGTGTATACCTATCATGGTGGGCTGGTTGATGCTGGGTAACTACTTCCAGCATATCCGACATCCGATCTTCGTAGCAGCGTACGCGGCTCTGGAGAGATCTATCTTTCAGATTATGTGCATCCTTGCGTATATAGCATTAGTCTTCAAAAATGATATTATTATATGCTTTAAAGCACTCGCCTGGCGAGGGTGGCATCTGTTGGCGCAGCTCTCTCTGCCCGTGATGCAGATACACTGGTGCATAGCGTTGGTCATCATTGGTCCATCGCCTCAGCCGGTTGCGGTTTCGGTCATTAACTTGCTACCGGTTAGCGTTACCACAGTCTGTTTCACGTATCCACTGGCATTGTTATTGACTGTACTGTTGGATATGGGACCAAAGGTAGGCAATCAATCAGTTAGATATGGGAGAAAAGGTAGGCAATCAATCAGTTAG
Protein
MLVSLCLLAALIEVKAADFNGEDYWLLPRLFDLDNYDECLSHPDGLYCLGSFHLTPAEQPNRVYELMTNYSKELYRFNRTVIRRGWCVSARESNVTAEPMNRFEHCAAEWARSRRMRARLLRLEYCRSHSDVHATRIPDKSELTFLIAIAVIILLNAASTAYDYATQNNEKKIALLTTWSVVANWRQLVEPYNSHRKSLQAIQGIKVIVSASICIVHSGMLLYTLFLYEPIEMEQVIANPLTMAVQNSTTVVHMFIVLSSFLTVHNLLKSSKPFGFGTFLRLLAKRIIRLSPVCLLVIGFAGTWWPRVSDGPLWPAAVAEQSKICREKFWAHAFFIHNLVEPDKFCLIPTWYIAVDMQMYILAVILTMILAKFSKKAVPILVILFIGATALNAGLAYIYEWKSLLYLAIVAENINSIFRDIPSLSHFYMSPWGSLPACLVGLIIGHLVHFTKGETFGNIYKRRLLSLYYVLCIPIMVGWLMLGNYFQHIRHPIFVAAYAALERSIFQIMCILAYIALVFKNDIIICFKALAWRGWHLLAQLSLPVMQIHWCIALVIIGPSPQPVAVSVINLLPVSVTTVCFTYPLALLLTVLLDMGPKVGNQSVRYGRKGRQSIS
Summary
Uniprot
A0A2A4K959
H9JU90
A0A2W1BIR4
A0A2H1WLB9
A0A2W1BKH9
A0A2A4K429
+ More
A0A212FM91 A0A2H1VFZ7 A0A0N0PCP5 A0A194Q086 A0A2A4K862 A0A0N1IH78 A0A2W1BRV2 A0A194Q6D5 A0A2A4K7P3 A0A212FMC6 A0A437AY35 A0A0L7KSK5 A0A2H1VKS3 A0A194Q1V4 A0A2H1WAT5 A0A2H1WCF8 A0A2A4JGF7 A0A212ETZ7 A0A2H1WAR4 A0A2W1BKB8 A0A212FGA7 A0A3S2P8D1 A0A2W1BK62 A0A0N1I4J5 A0A2H1WCA0 A0A194QFD0 A0A0N0PAP1 M4M6W5 A0A194QF68 A0A0N1IBJ6 A0A2A4JGW2 A0A2W1BKH3 A0A1Y1M2Q0 A0A182YEG1 B0WNX6 B3MGQ2 H9JW27 A0A1S4GUU3 A0A182I2D7 Q7QIT8 A0A182KQ82 A0A0N0PEC2 A0A182MV59 A0A182WWU1 Q7PTG8 A0A182FHD9 A0A182NJC1 A0A182R4J9 A0A182VP89 A0A182PUZ7 H9J531 A1Z6Q3 A0A084WED5 Q16PV8 A0A182R1B6 A0A2J7Q6G4 A0A1Y1KVX9 A0A1S4FTS9 W5JFI4 A0A182UXI0 A0A1W4VFG7 A0A182KD11 B5E1C2 A0A1W4V1V5 A0A182YNQ4 A0A182TKI8 A0A182JNH6 A0A0N0P9S1
A0A212FM91 A0A2H1VFZ7 A0A0N0PCP5 A0A194Q086 A0A2A4K862 A0A0N1IH78 A0A2W1BRV2 A0A194Q6D5 A0A2A4K7P3 A0A212FMC6 A0A437AY35 A0A0L7KSK5 A0A2H1VKS3 A0A194Q1V4 A0A2H1WAT5 A0A2H1WCF8 A0A2A4JGF7 A0A212ETZ7 A0A2H1WAR4 A0A2W1BKB8 A0A212FGA7 A0A3S2P8D1 A0A2W1BK62 A0A0N1I4J5 A0A2H1WCA0 A0A194QFD0 A0A0N0PAP1 M4M6W5 A0A194QF68 A0A0N1IBJ6 A0A2A4JGW2 A0A2W1BKH3 A0A1Y1M2Q0 A0A182YEG1 B0WNX6 B3MGQ2 H9JW27 A0A1S4GUU3 A0A182I2D7 Q7QIT8 A0A182KQ82 A0A0N0PEC2 A0A182MV59 A0A182WWU1 Q7PTG8 A0A182FHD9 A0A182NJC1 A0A182R4J9 A0A182VP89 A0A182PUZ7 H9J531 A1Z6Q3 A0A084WED5 Q16PV8 A0A182R1B6 A0A2J7Q6G4 A0A1Y1KVX9 A0A1S4FTS9 W5JFI4 A0A182UXI0 A0A1W4VFG7 A0A182KD11 B5E1C2 A0A1W4V1V5 A0A182YNQ4 A0A182TKI8 A0A182JNH6 A0A0N0P9S1
Pubmed
EMBL
NWSH01000045
PCG80260.1
BABH01004270
BABH01004271
KZ150097
PZC73595.1
+ More
ODYU01009435 SOQ53859.1 KZ149997 PZC75398.1 NWSH01000169 PCG78819.1 AGBW02007654 OWR54851.1 ODYU01002360 SOQ39759.1 KQ460635 KPJ13420.1 KQ459582 KPI98971.1 PCG80259.1 KPJ13421.1 PZC75396.1 KPI98970.1 PCG80257.1 OWR54850.1 RSAL01000280 RVE43059.1 JTDY01006392 KOB66051.1 ODYU01003108 SOQ41449.1 KPI98974.1 ODYU01007420 SOQ50170.1 SOQ50174.1 NWSH01001667 PCG70472.1 AGBW02012502 OWR44975.1 SOQ50171.1 KZ150068 PZC74094.1 AGBW02008687 OWR52781.1 RSAL01000205 RVE44373.1 PZC74095.1 KQ459449 KPJ00879.1 ODYU01007576 SOQ50462.1 KQ459053 KPJ04119.1 KQ461184 KPJ07529.1 JX846987 AGG55004.1 KPJ04127.1 KPJ07525.1 NWSH01001633 PCG70632.1 KZ150085 PZC73777.1 GEZM01042633 JAV79751.1 DS232017 EDS31995.1 CH902619 EDV36810.2 BABH01025581 AAAB01008807 APCN01000146 EAA04081.4 KQ459900 KPJ19352.1 AXCM01001275 EAA03947.5 BABH01038834 AE013599 AAF57404.4 ATLV01023202 ATLV01023203 ATLV01023204 KE525341 KFB48579.1 CH477771 EAT36390.1 AXCN02000448 AXCN02000449 NEVH01017478 PNF24171.1 GEZM01074205 GEZM01074204 JAV64641.1 ADMH02001642 ETN61640.1 CM000071 EDY68727.2 KPJ00884.1
ODYU01009435 SOQ53859.1 KZ149997 PZC75398.1 NWSH01000169 PCG78819.1 AGBW02007654 OWR54851.1 ODYU01002360 SOQ39759.1 KQ460635 KPJ13420.1 KQ459582 KPI98971.1 PCG80259.1 KPJ13421.1 PZC75396.1 KPI98970.1 PCG80257.1 OWR54850.1 RSAL01000280 RVE43059.1 JTDY01006392 KOB66051.1 ODYU01003108 SOQ41449.1 KPI98974.1 ODYU01007420 SOQ50170.1 SOQ50174.1 NWSH01001667 PCG70472.1 AGBW02012502 OWR44975.1 SOQ50171.1 KZ150068 PZC74094.1 AGBW02008687 OWR52781.1 RSAL01000205 RVE44373.1 PZC74095.1 KQ459449 KPJ00879.1 ODYU01007576 SOQ50462.1 KQ459053 KPJ04119.1 KQ461184 KPJ07529.1 JX846987 AGG55004.1 KPJ04127.1 KPJ07525.1 NWSH01001633 PCG70632.1 KZ150085 PZC73777.1 GEZM01042633 JAV79751.1 DS232017 EDS31995.1 CH902619 EDV36810.2 BABH01025581 AAAB01008807 APCN01000146 EAA04081.4 KQ459900 KPJ19352.1 AXCM01001275 EAA03947.5 BABH01038834 AE013599 AAF57404.4 ATLV01023202 ATLV01023203 ATLV01023204 KE525341 KFB48579.1 CH477771 EAT36390.1 AXCN02000448 AXCN02000449 NEVH01017478 PNF24171.1 GEZM01074205 GEZM01074204 JAV64641.1 ADMH02001642 ETN61640.1 CM000071 EDY68727.2 KPJ00884.1
Proteomes
UP000218220
UP000005204
UP000007151
UP000053240
UP000053268
UP000283053
+ More
UP000037510 UP000076408 UP000002320 UP000007801 UP000075840 UP000007062 UP000075882 UP000075883 UP000076407 UP000069272 UP000075884 UP000075900 UP000075903 UP000075885 UP000000803 UP000030765 UP000008820 UP000075886 UP000235965 UP000000673 UP000192221 UP000075881 UP000001819 UP000075902
UP000037510 UP000076408 UP000002320 UP000007801 UP000075840 UP000007062 UP000075882 UP000075883 UP000076407 UP000069272 UP000075884 UP000075900 UP000075903 UP000075885 UP000000803 UP000030765 UP000008820 UP000075886 UP000235965 UP000000673 UP000192221 UP000075881 UP000001819 UP000075902
Interpro
ProteinModelPortal
A0A2A4K959
H9JU90
A0A2W1BIR4
A0A2H1WLB9
A0A2W1BKH9
A0A2A4K429
+ More
A0A212FM91 A0A2H1VFZ7 A0A0N0PCP5 A0A194Q086 A0A2A4K862 A0A0N1IH78 A0A2W1BRV2 A0A194Q6D5 A0A2A4K7P3 A0A212FMC6 A0A437AY35 A0A0L7KSK5 A0A2H1VKS3 A0A194Q1V4 A0A2H1WAT5 A0A2H1WCF8 A0A2A4JGF7 A0A212ETZ7 A0A2H1WAR4 A0A2W1BKB8 A0A212FGA7 A0A3S2P8D1 A0A2W1BK62 A0A0N1I4J5 A0A2H1WCA0 A0A194QFD0 A0A0N0PAP1 M4M6W5 A0A194QF68 A0A0N1IBJ6 A0A2A4JGW2 A0A2W1BKH3 A0A1Y1M2Q0 A0A182YEG1 B0WNX6 B3MGQ2 H9JW27 A0A1S4GUU3 A0A182I2D7 Q7QIT8 A0A182KQ82 A0A0N0PEC2 A0A182MV59 A0A182WWU1 Q7PTG8 A0A182FHD9 A0A182NJC1 A0A182R4J9 A0A182VP89 A0A182PUZ7 H9J531 A1Z6Q3 A0A084WED5 Q16PV8 A0A182R1B6 A0A2J7Q6G4 A0A1Y1KVX9 A0A1S4FTS9 W5JFI4 A0A182UXI0 A0A1W4VFG7 A0A182KD11 B5E1C2 A0A1W4V1V5 A0A182YNQ4 A0A182TKI8 A0A182JNH6 A0A0N0P9S1
A0A212FM91 A0A2H1VFZ7 A0A0N0PCP5 A0A194Q086 A0A2A4K862 A0A0N1IH78 A0A2W1BRV2 A0A194Q6D5 A0A2A4K7P3 A0A212FMC6 A0A437AY35 A0A0L7KSK5 A0A2H1VKS3 A0A194Q1V4 A0A2H1WAT5 A0A2H1WCF8 A0A2A4JGF7 A0A212ETZ7 A0A2H1WAR4 A0A2W1BKB8 A0A212FGA7 A0A3S2P8D1 A0A2W1BK62 A0A0N1I4J5 A0A2H1WCA0 A0A194QFD0 A0A0N0PAP1 M4M6W5 A0A194QF68 A0A0N1IBJ6 A0A2A4JGW2 A0A2W1BKH3 A0A1Y1M2Q0 A0A182YEG1 B0WNX6 B3MGQ2 H9JW27 A0A1S4GUU3 A0A182I2D7 Q7QIT8 A0A182KQ82 A0A0N0PEC2 A0A182MV59 A0A182WWU1 Q7PTG8 A0A182FHD9 A0A182NJC1 A0A182R4J9 A0A182VP89 A0A182PUZ7 H9J531 A1Z6Q3 A0A084WED5 Q16PV8 A0A182R1B6 A0A2J7Q6G4 A0A1Y1KVX9 A0A1S4FTS9 W5JFI4 A0A182UXI0 A0A1W4VFG7 A0A182KD11 B5E1C2 A0A1W4V1V5 A0A182YNQ4 A0A182TKI8 A0A182JNH6 A0A0N0P9S1
Ontologies
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.994234
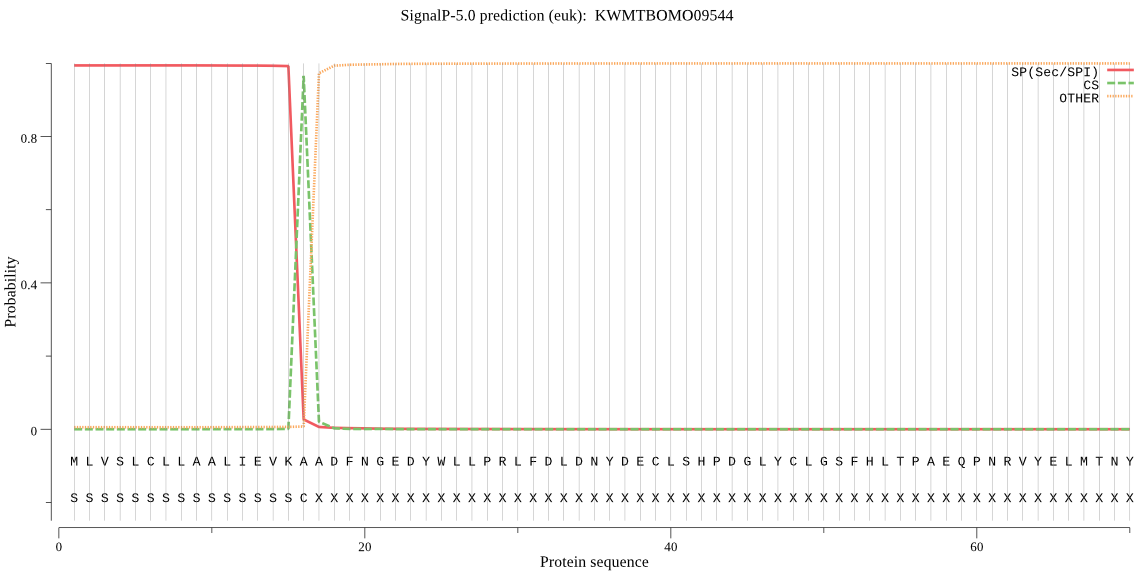
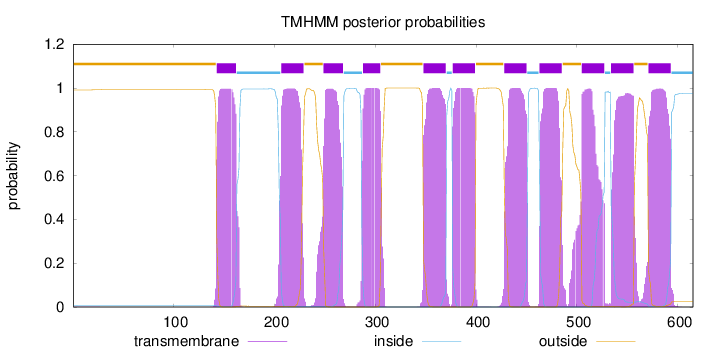
Length:
615
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
238.94591
Exp number, first 60 AAs:
0.02249
Total prob of N-in:
0.00859
outside
1 - 142
TMhelix
143 - 162
inside
163 - 206
TMhelix
207 - 229
outside
230 - 248
TMhelix
249 - 268
inside
269 - 287
TMhelix
288 - 305
outside
306 - 347
TMhelix
348 - 370
inside
371 - 376
TMhelix
377 - 399
outside
400 - 427
TMhelix
428 - 450
inside
451 - 462
TMhelix
463 - 485
outside
486 - 504
TMhelix
505 - 527
inside
528 - 533
TMhelix
534 - 556
outside
557 - 570
TMhelix
571 - 593
inside
594 - 615
Population Genetic Test Statistics
Pi
181.02389
Theta
173.522513
Tajima's D
0.758003
CLR
0.433525
CSRT
0.592070396480176
Interpretation
Uncertain
