Pre Gene Modal
BGIBMGA013027
Annotation
PREDICTED:_phagocyte_signaling-impaired_protein_isoform_X1_[Bombyx_mori]
Full name
Phagocyte signaling-impaired protein
Alternative Name
N-terminal acetyltransferase B complex subunit MDM20 homolog
N-terminal acetyltransferase B complex subunit NAA25 homolog
N-terminal acetyltransferase B complex subunit NAA25 homolog
Location in the cell
Cytoplasmic Reliability : 1.342 Nuclear Reliability : 1.292
Sequence
CDS
ATGGCGTTAAGATCGCAGCATGTGCACGATGGTGGCGTTGTAGAACGACGGTTGCGGCCTGTATATGATTGGCTGGATAATGGGTATAATAAAAAGGCTTTACAAGAGGCGGAAAAGGTATTAAAAAAGAGTCCATCGTTACAAGCGGCGCGAGCTTTGAAAGCTCTAGCACTTTATAGGTTAGGTAGAGCCCAAGAGGCAGGAGCTGTGCTAGATGCATTGGTCGAGGAGAAGCCTAGCGATGACACGACATTACAGGCGATGACAATGTCATTTCGAGAGTCTCAGCAATTGCACAAAGTTTGTACCATCTATGAAAATGCTGTGAAAATAGATACAACAAATGAGGAACTACATTCTCACCTGTTTATGTCATATGTACGGGTCGGAGATTTTCGATCGCAGCAACGAGCAGCAATGGCAATATACAAGTTTGCGCCAAAAAATCCATATTATTTCTGGGCAGTCATGAGCATAGTTTTGCAGGCCAAAGAAACAGAAGACACAACAAAAAGGAACATTCTCCTCACTCTTGCCCAAAGAATGATAGACAATTTTATATCTGAAAATCGAATGGAAGCCGAACAAGAGGCCCGCCTGTACATAATGATATTAGAACTACAAGAGAAATGGGAAGATATAATCAAATTTATAGAAAGCCCTTTGTATGGACAACTTATACCTGGTTCTATATCGCAAGCTTGTATTCCGTATTTGAAAAAGTTGGGTAGATGGAAAAAAGTCAACATCCTCTGTAAAGAGCTACTATATGATAATTTAGATAGATGGGATTATTACATGCCTTATTTCGACTCTGTCTTCCATTTGATGAAAGAGAATAGCAACGAAGAAAATATAGACGATACGGCTGAGAAATGCCATGAATTTATATGTCAGTTAGTCGAAAGTATGTCTTCCGGGCGAACATTGAGAGGTCCTTACTTAGCCAGACTGGAATTGTGGAAACGTTTGTCTGTCGAAGGAGATCCGACAGCGTTACTCGGTAGTGCGTTGGCACTCTGCGTGCAGTATTTAAGGGTGTTTGCAAGTAAACCTTGTGCTGTACCAGACTTGAAAGTTTATTTAGCGATGATACCCCAAGAAGAAAGGGAGGCTAACTGCAGAGATTTCCTGACATGCCTCGGTTTCGATGAAAACTCTGAACCAGAAACGATGGACGACATACAGCGTCACATCTCGTGTCTGACAGCATGGCGCCTGACAGCCCCTCGTCCTGGTACGACAGACGCCCTCCAGCTAGCCGCTACGCTCAGGAGGCATTACCTCAGGTCCCTCGAGAAAAACATGATCAAGTACTCAACTACTGAGTTCTGTTCCGCAGATAGCTATGCGATACTAGCGGCTCATCATTATTTTTATGCTGCAACAGAACAGAAGAGCTCAAAACCCATCCTCGAAGCCTTGTGTTTGTTAGAATTAGTATTGTTGTATTCACCAGCGAATTATCACGTGAAGCTTCTTCTAGTGCAACTGTATCATCTGCTAGGTAACGCATTAGCGGCAGATTCGATCTACCGTCGCCTAGAGCTGAAGCACATCCAGCTGGTGTCGCTGGGCTGGCTGCACACTGCGCGAGCGGACGCGGCCTGCGTGCCGACGCGCGCCCTGCAGCTGCTGGCCGACCTTAGGGCCTTCCACACGCACCACGCTAAGGATAGCATCGAGCACATGACGTACGCGTACAAGTTCGGCACGTTCGAGAAGGTGGTGGAGCTGCGCGCGTGGGCGGCGCGGCTGGAGGCGTGCGCGCTCTGTGCCGTGTCGGCGCGCCGCCGCGACCTGCTGGCGCTGCTCGCCGCCCCCGCGCCGCCCGCGCCCCCCGCGCCCCTGCACGCCGCGCACACCGACAACCGCGACCTGAACATCATCGTGAACTGGGAGCCGCCCGAGCTGCAGGATCCTGATTTGAGATCTCGAACGTTCGATCAGGATATGGCCTACCTCCGCTTGACGGACGCGCTGCTGTCCTCGGTCACGCTGTGCACGGAACTCGCCGAGAACAAACCGATCGAAGAGAAGATGCAACATTACGAGCAACTGCGAACCTGCGTCGACACGTTCAGCGAAGCGATGGAGAAATGTACAGAGAAATATTGCGTTTACGAGAAGATCACTATATCCGGACCGTTTCCTTCCAGGATAATTGGTTTCGTAAGTTCGCCGGTGCCGTATCGGGAACTGTACTCGATAACGCTGCGCATGATCGGCGAGCTGTCCATGAACAACACTGACAAAGTGCACGAGCTGTGCGATCAAGTCGTCGCCATCATTTTGATTTCGTCAAAACTTATCGTCCAAGAGATCCGATACGGCGCTGAAGACAACGGCTGGTCGATGCGGGATAGACTCGAGTCGCTGTCCAATTATTTACAATCAATCGGCGTGCTAACGTATTTACTGGGCGTTTGCTACGAGATGATAACTCCGACGAACACAAAGAAATCTAAGAAGAAAGCGAACCGCTCCGTCGATCAGATGCAAACGATAAAATTAATTGACAACATAAACGTGACGTTGCAACAGGCGATCGCTTCGCTCGAGGAGCTCATGGACCTCTGGCCTGACTACGACTTCTCGGTCGACCTGGAAGAAGAACTGACCAGAATGAACATTAACCCAACCGCACCCAGCCCGGTCGAGAATAAACTGAAGCTAGGCCTCGACAAGATAATTGAAGACATACAGAATATACTGAAACGAAAGACTGCTTATTTGAAAGGCTTAATACTAAATACTAATACTTAA
Protein
MALRSQHVHDGGVVERRLRPVYDWLDNGYNKKALQEAEKVLKKSPSLQAARALKALALYRLGRAQEAGAVLDALVEEKPSDDTTLQAMTMSFRESQQLHKVCTIYENAVKIDTTNEELHSHLFMSYVRVGDFRSQQRAAMAIYKFAPKNPYYFWAVMSIVLQAKETEDTTKRNILLTLAQRMIDNFISENRMEAEQEARLYIMILELQEKWEDIIKFIESPLYGQLIPGSISQACIPYLKKLGRWKKVNILCKELLYDNLDRWDYYMPYFDSVFHLMKENSNEENIDDTAEKCHEFICQLVESMSSGRTLRGPYLARLELWKRLSVEGDPTALLGSALALCVQYLRVFASKPCAVPDLKVYLAMIPQEEREANCRDFLTCLGFDENSEPETMDDIQRHISCLTAWRLTAPRPGTTDALQLAATLRRHYLRSLEKNMIKYSTTEFCSADSYAILAAHHYFYAATEQKSSKPILEALCLLELVLLYSPANYHVKLLLVQLYHLLGNALAADSIYRRLELKHIQLVSLGWLHTARADAACVPTRALQLLADLRAFHTHHAKDSIEHMTYAYKFGTFEKVVELRAWAARLEACALCAVSARRRDLLALLAAPAPPAPPAPLHAAHTDNRDLNIIVNWEPPELQDPDLRSRTFDQDMAYLRLTDALLSSVTLCTELAENKPIEEKMQHYEQLRTCVDTFSEAMEKCTEKYCVYEKITISGPFPSRIIGFVSSPVPYRELYSITLRMIGELSMNNTDKVHELCDQVVAIILISSKLIVQEIRYGAEDNGWSMRDRLESLSNYLQSIGVLTYLLGVCYEMITPTNTKKSKKKANRSVDQMQTIKLIDNINVTLQQAIASLEELMDLWPDYDFSVDLEEELTRMNINPTAPSPVENKLKLGLDKIIEDIQNILKRKTAYLKGLILNTNT
Summary
Description
Non-catalytic subunit of the NatB complex which catalyzes acetylation of the N-terminal methionine residues of proteins beginning with Met-Asp or Met-Glu (By similarity). Has 2 roles in the larval immune response: required both for the phagocytic degradation of internalized bacteria and for the induction of Defensin in the fat body. Within the phagocytic blood cells, has a role in detection of infection and activation of the humoral immune response (By similarity).
Non-catalytic subunit of the NatB complex which catalyzes acetylation of the N-terminal methionine residues of proteins beginning with Met-Asp or Met-Glu (By similarity). Has 2 roles in the larval immune response: required both for the phagocytic degradation of internalized bacteria and for the induction of Defensin in the fat body. Within the phagocytic blood cells, has a role in detection of infection and activation of the humoral immune response.
Non-catalytic subunit of the NatB complex which catalyzes acetylation of the N-terminal methionine residues of proteins beginning with Met-Asp or Met-Glu (By similarity). Has 2 roles in the larval immune response: required both for the phagocytic degradation of internalized bacteria and for the induction of Defensin in the fat body. Within the phagocytic blood cells, has a role in detection of infection and activation of the humoral immune response.
Subunit
Component of the N-terminal acetyltransferase B (NatB) complex.
Similarity
Belongs to the MDM20/NAA25 family.
Keywords
Antimicrobial
Coiled coil
Complete proteome
Immunity
Innate immunity
Lysosome
Reference proteome
TPR repeat
Repeat
Feature
chain Phagocyte signaling-impaired protein
Uniprot
A0A2W1BFN4
A0A437BBZ6
S4PGM8
A0A2A4K4H8
A0A212FMR7
A0A194Q1A4
+ More
A0A1Y1NI51 A0A1Y1NI62 A0A1W4XH84 A0A1W4XSN3 D2A4T4 A0A1W4XSU1 A0A2J7RM85 U4UXH7 A0A1L8E649 A0A1B0ESZ5 N6T4C0 A0A0K8VWE8 E0VJ79 A0A034VSP0 A0A0L0C3W8 W8AQU8 A0A0A1WNR0 A0A1A9VB89 A0A1I8P4E4 A0A1B0G7Y7 A0A1A9ZHL7 A0A1A9WJA3 A0A1A9XGA6 A0A1B0BNK7 A0A336LYJ4 B3LXU9 A0A067RFA8 A0A1B6CP03 T1PK04 A0A1I8NGJ5 B0WRR7 A0A1Q3F5B7 B4KE33 B4NGC4 A0A084VS51 B4M4U1 A0A0P4WVZ3 A0A0N7ZEZ1 A0A0N8AKT1 A0A1S4F6R8 A0A182RXJ7 B3NZ24 Q7PYI4 A0A0P5Y900 A0A0P6AKF4 A0A0P5WJQ9 A0A0P4ZJY0 A0A182XBG5 A0A164YBF2 A0A182N4B9 A0A0P4WPL9 A0A0P5Y5B3 A0A182KLX7 B4PP53 A0A0P5V9L5 A0A182I0A5 A0A182GT94 Q17DK2 W5JG94 A0A182F881 A0A2M4AKV0 A0A0M4EVG2 A0A182TUQ7 A0A182VR33 A0A0P5QDG2 A0A0K8V4Q5 A0A182VIG1 A0A182JR70 A0A2M4BC68 A0A2M4BCA6 A0A0N0U4G9 A0A0P5R8Z7 A0A182M2T0 U3Q053 Q9VDQ7 A0A0P5Z993 Q294E0 B4GL86 A0A182J0P0 A0A026WNM2 A0A3L8E3P5 B4JSZ1 E2BAH2
A0A1Y1NI51 A0A1Y1NI62 A0A1W4XH84 A0A1W4XSN3 D2A4T4 A0A1W4XSU1 A0A2J7RM85 U4UXH7 A0A1L8E649 A0A1B0ESZ5 N6T4C0 A0A0K8VWE8 E0VJ79 A0A034VSP0 A0A0L0C3W8 W8AQU8 A0A0A1WNR0 A0A1A9VB89 A0A1I8P4E4 A0A1B0G7Y7 A0A1A9ZHL7 A0A1A9WJA3 A0A1A9XGA6 A0A1B0BNK7 A0A336LYJ4 B3LXU9 A0A067RFA8 A0A1B6CP03 T1PK04 A0A1I8NGJ5 B0WRR7 A0A1Q3F5B7 B4KE33 B4NGC4 A0A084VS51 B4M4U1 A0A0P4WVZ3 A0A0N7ZEZ1 A0A0N8AKT1 A0A1S4F6R8 A0A182RXJ7 B3NZ24 Q7PYI4 A0A0P5Y900 A0A0P6AKF4 A0A0P5WJQ9 A0A0P4ZJY0 A0A182XBG5 A0A164YBF2 A0A182N4B9 A0A0P4WPL9 A0A0P5Y5B3 A0A182KLX7 B4PP53 A0A0P5V9L5 A0A182I0A5 A0A182GT94 Q17DK2 W5JG94 A0A182F881 A0A2M4AKV0 A0A0M4EVG2 A0A182TUQ7 A0A182VR33 A0A0P5QDG2 A0A0K8V4Q5 A0A182VIG1 A0A182JR70 A0A2M4BC68 A0A2M4BCA6 A0A0N0U4G9 A0A0P5R8Z7 A0A182M2T0 U3Q053 Q9VDQ7 A0A0P5Z993 Q294E0 B4GL86 A0A182J0P0 A0A026WNM2 A0A3L8E3P5 B4JSZ1 E2BAH2
Pubmed
EMBL
KZ150097
PZC73598.1
RSAL01000093
RVE47920.1
GAIX01003592
JAA88968.1
+ More
NWSH01000169 PCG78814.1 AGBW02007651 OWR55035.1 KQ459582 KPI99103.1 GEZM01001684 GEZM01001682 JAV97582.1 GEZM01001686 GEZM01001681 JAV97583.1 KQ971344 EFA05265.1 NEVH01002568 PNF41941.1 KB632404 ERL95051.1 GFDF01000092 JAV13992.1 AJWK01005336 AJWK01005337 AJWK01005338 AJWK01005339 AJWK01005340 APGK01052815 KB741216 ENN72468.1 GDHF01009102 JAI43212.1 DS235221 EEB13435.1 GAKP01012641 JAC46311.1 JRES01000944 KNC26947.1 GAMC01018143 JAB88412.1 GBXI01013815 GBXI01005535 JAD00477.1 JAD08757.1 CCAG010010579 JXJN01017451 JXJN01017452 UFQT01000042 SSX18738.1 CH902617 EDV41756.1 KK852507 KDR22452.1 GEDC01022197 JAS15101.1 KA648293 AFP62922.1 DS232059 EDS33476.1 GFDL01012293 JAV22752.1 CH933806 EDW16054.2 CH964251 EDW83341.1 ATLV01015810 KE525036 KFB40795.1 CH940652 EDW59652.1 GDIP01251815 JAI71586.1 GDIP01252009 JAI71392.1 GDIQ01266011 GDIQ01006869 JAJ85713.1 CH954181 EDV48427.1 AAAB01008987 GDIP01062739 JAM40976.1 GDIP01028144 JAM75571.1 GDIP01085477 JAM18238.1 GDIP01224864 JAI98537.1 LRGB01000930 KZS15079.1 GDIP01254711 JAI68690.1 GDIP01062740 JAM40975.1 CM000160 EDW96092.2 GDIP01118381 JAL85333.1 APCN01002057 JXUM01086749 JXUM01086750 JXUM01086751 KQ563588 KXJ73627.1 CH477293 EAT44499.1 ADMH02001488 ETN62358.1 GGFK01008089 MBW41410.1 CP012526 ALC47317.1 GDIQ01217141 GDIQ01215944 GDIQ01214335 GDIQ01139469 GDIQ01137013 GDIQ01112469 GDIQ01110770 GDIP01079779 GDIQ01041960 JAL14713.1 JAM23936.1 GDHF01024171 GDHF01018425 JAI28143.1 JAI33889.1 GGFJ01001471 MBW50612.1 GGFJ01001470 MBW50611.1 KQ435833 KOX71671.1 GDIQ01108930 JAL42796.1 AXCM01003379 BT150356 AGW52165.1 AE014297 BT010121 AAQ22590.1 GDIP01047043 JAM56672.1 CM000070 CH479185 EDW38310.1 KK107159 EZA56704.1 QOIP01000001 RLU27025.1 CH916373 EDV94881.1 GL446730 EFN87304.1
NWSH01000169 PCG78814.1 AGBW02007651 OWR55035.1 KQ459582 KPI99103.1 GEZM01001684 GEZM01001682 JAV97582.1 GEZM01001686 GEZM01001681 JAV97583.1 KQ971344 EFA05265.1 NEVH01002568 PNF41941.1 KB632404 ERL95051.1 GFDF01000092 JAV13992.1 AJWK01005336 AJWK01005337 AJWK01005338 AJWK01005339 AJWK01005340 APGK01052815 KB741216 ENN72468.1 GDHF01009102 JAI43212.1 DS235221 EEB13435.1 GAKP01012641 JAC46311.1 JRES01000944 KNC26947.1 GAMC01018143 JAB88412.1 GBXI01013815 GBXI01005535 JAD00477.1 JAD08757.1 CCAG010010579 JXJN01017451 JXJN01017452 UFQT01000042 SSX18738.1 CH902617 EDV41756.1 KK852507 KDR22452.1 GEDC01022197 JAS15101.1 KA648293 AFP62922.1 DS232059 EDS33476.1 GFDL01012293 JAV22752.1 CH933806 EDW16054.2 CH964251 EDW83341.1 ATLV01015810 KE525036 KFB40795.1 CH940652 EDW59652.1 GDIP01251815 JAI71586.1 GDIP01252009 JAI71392.1 GDIQ01266011 GDIQ01006869 JAJ85713.1 CH954181 EDV48427.1 AAAB01008987 GDIP01062739 JAM40976.1 GDIP01028144 JAM75571.1 GDIP01085477 JAM18238.1 GDIP01224864 JAI98537.1 LRGB01000930 KZS15079.1 GDIP01254711 JAI68690.1 GDIP01062740 JAM40975.1 CM000160 EDW96092.2 GDIP01118381 JAL85333.1 APCN01002057 JXUM01086749 JXUM01086750 JXUM01086751 KQ563588 KXJ73627.1 CH477293 EAT44499.1 ADMH02001488 ETN62358.1 GGFK01008089 MBW41410.1 CP012526 ALC47317.1 GDIQ01217141 GDIQ01215944 GDIQ01214335 GDIQ01139469 GDIQ01137013 GDIQ01112469 GDIQ01110770 GDIP01079779 GDIQ01041960 JAL14713.1 JAM23936.1 GDHF01024171 GDHF01018425 JAI28143.1 JAI33889.1 GGFJ01001471 MBW50612.1 GGFJ01001470 MBW50611.1 KQ435833 KOX71671.1 GDIQ01108930 JAL42796.1 AXCM01003379 BT150356 AGW52165.1 AE014297 BT010121 AAQ22590.1 GDIP01047043 JAM56672.1 CM000070 CH479185 EDW38310.1 KK107159 EZA56704.1 QOIP01000001 RLU27025.1 CH916373 EDV94881.1 GL446730 EFN87304.1
Proteomes
UP000283053
UP000218220
UP000007151
UP000053268
UP000192223
UP000007266
+ More
UP000235965 UP000030742 UP000092461 UP000019118 UP000009046 UP000037069 UP000078200 UP000095300 UP000092444 UP000092445 UP000091820 UP000092443 UP000092460 UP000007801 UP000027135 UP000095301 UP000002320 UP000009192 UP000007798 UP000030765 UP000008792 UP000075900 UP000008711 UP000007062 UP000076407 UP000076858 UP000075884 UP000075882 UP000002282 UP000075840 UP000069940 UP000249989 UP000008820 UP000000673 UP000069272 UP000092553 UP000075902 UP000075920 UP000075903 UP000075881 UP000053105 UP000075883 UP000000803 UP000001819 UP000008744 UP000075880 UP000053097 UP000279307 UP000001070 UP000008237
UP000235965 UP000030742 UP000092461 UP000019118 UP000009046 UP000037069 UP000078200 UP000095300 UP000092444 UP000092445 UP000091820 UP000092443 UP000092460 UP000007801 UP000027135 UP000095301 UP000002320 UP000009192 UP000007798 UP000030765 UP000008792 UP000075900 UP000008711 UP000007062 UP000076407 UP000076858 UP000075884 UP000075882 UP000002282 UP000075840 UP000069940 UP000249989 UP000008820 UP000000673 UP000069272 UP000092553 UP000075902 UP000075920 UP000075903 UP000075881 UP000053105 UP000075883 UP000000803 UP000001819 UP000008744 UP000075880 UP000053097 UP000279307 UP000001070 UP000008237
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BFN4
A0A437BBZ6
S4PGM8
A0A2A4K4H8
A0A212FMR7
A0A194Q1A4
+ More
A0A1Y1NI51 A0A1Y1NI62 A0A1W4XH84 A0A1W4XSN3 D2A4T4 A0A1W4XSU1 A0A2J7RM85 U4UXH7 A0A1L8E649 A0A1B0ESZ5 N6T4C0 A0A0K8VWE8 E0VJ79 A0A034VSP0 A0A0L0C3W8 W8AQU8 A0A0A1WNR0 A0A1A9VB89 A0A1I8P4E4 A0A1B0G7Y7 A0A1A9ZHL7 A0A1A9WJA3 A0A1A9XGA6 A0A1B0BNK7 A0A336LYJ4 B3LXU9 A0A067RFA8 A0A1B6CP03 T1PK04 A0A1I8NGJ5 B0WRR7 A0A1Q3F5B7 B4KE33 B4NGC4 A0A084VS51 B4M4U1 A0A0P4WVZ3 A0A0N7ZEZ1 A0A0N8AKT1 A0A1S4F6R8 A0A182RXJ7 B3NZ24 Q7PYI4 A0A0P5Y900 A0A0P6AKF4 A0A0P5WJQ9 A0A0P4ZJY0 A0A182XBG5 A0A164YBF2 A0A182N4B9 A0A0P4WPL9 A0A0P5Y5B3 A0A182KLX7 B4PP53 A0A0P5V9L5 A0A182I0A5 A0A182GT94 Q17DK2 W5JG94 A0A182F881 A0A2M4AKV0 A0A0M4EVG2 A0A182TUQ7 A0A182VR33 A0A0P5QDG2 A0A0K8V4Q5 A0A182VIG1 A0A182JR70 A0A2M4BC68 A0A2M4BCA6 A0A0N0U4G9 A0A0P5R8Z7 A0A182M2T0 U3Q053 Q9VDQ7 A0A0P5Z993 Q294E0 B4GL86 A0A182J0P0 A0A026WNM2 A0A3L8E3P5 B4JSZ1 E2BAH2
A0A1Y1NI51 A0A1Y1NI62 A0A1W4XH84 A0A1W4XSN3 D2A4T4 A0A1W4XSU1 A0A2J7RM85 U4UXH7 A0A1L8E649 A0A1B0ESZ5 N6T4C0 A0A0K8VWE8 E0VJ79 A0A034VSP0 A0A0L0C3W8 W8AQU8 A0A0A1WNR0 A0A1A9VB89 A0A1I8P4E4 A0A1B0G7Y7 A0A1A9ZHL7 A0A1A9WJA3 A0A1A9XGA6 A0A1B0BNK7 A0A336LYJ4 B3LXU9 A0A067RFA8 A0A1B6CP03 T1PK04 A0A1I8NGJ5 B0WRR7 A0A1Q3F5B7 B4KE33 B4NGC4 A0A084VS51 B4M4U1 A0A0P4WVZ3 A0A0N7ZEZ1 A0A0N8AKT1 A0A1S4F6R8 A0A182RXJ7 B3NZ24 Q7PYI4 A0A0P5Y900 A0A0P6AKF4 A0A0P5WJQ9 A0A0P4ZJY0 A0A182XBG5 A0A164YBF2 A0A182N4B9 A0A0P4WPL9 A0A0P5Y5B3 A0A182KLX7 B4PP53 A0A0P5V9L5 A0A182I0A5 A0A182GT94 Q17DK2 W5JG94 A0A182F881 A0A2M4AKV0 A0A0M4EVG2 A0A182TUQ7 A0A182VR33 A0A0P5QDG2 A0A0K8V4Q5 A0A182VIG1 A0A182JR70 A0A2M4BC68 A0A2M4BCA6 A0A0N0U4G9 A0A0P5R8Z7 A0A182M2T0 U3Q053 Q9VDQ7 A0A0P5Z993 Q294E0 B4GL86 A0A182J0P0 A0A026WNM2 A0A3L8E3P5 B4JSZ1 E2BAH2
PDB
5K18
E-value=0.0096999,
Score=95
Ontologies
GO
PANTHER
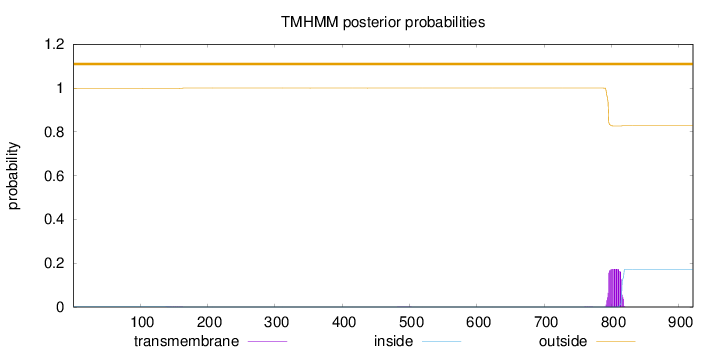
Topology
Subcellular location
Lysosome
Blood cell lysosomes. With evidence from 2 publications.
Length:
921
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.67072
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00138
outside
1 - 921
Population Genetic Test Statistics
Pi
211.703113
Theta
193.00704
Tajima's D
-1.066539
CLR
2.966829
CSRT
0.12364381780911
Interpretation
Uncertain
