Gene
KWMTBOMO09531 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005786
Annotation
myo-inositol_oxygenase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.556
Sequence
CDS
ATGGCCGCCGAAGTTGAAAAGAAGCCTGTGTCCATGATCGACCCGTCGCAGCTTCTGCGTCCAGAGCCTACGTTCAATGACAAGCCCATCGACGCGTTCAGGGACTACAGCGTCGACGACACGGACCCCATCAAGGAACGGGTCCGACGCACTTACTACTTGATGCACTCCAATGTTACCGTCGATTTGGTCAAACAAAAACGCGAGAAATGGCTAAAGTTCAACCACTTCAAGGCCACAGTGAAGGATGCTCTCATCAAGCTCAATGAGCTGGTGGACGAGTCAGACCCCGACACGGACCTACCAAACATCGTGCACGCTTTCCAGACCGCTGAAAGGATCAGAGAGGATCACCCTGATGAGGACTGGTTCCACCTTACTGGCCTCATTCATGATTTAGGCAAGGTAATGGCATTTTATGACGAGCCGCAGTGGTGCGTGGTCGGAGACACTTTCCCCGTTGGTTGTAAATGGGCTGACTCCATCGTCTATGGTCCCGAGAGCTTCAAGGACAACCCCGACACTTATAACCCTAAATACAACACCAAATATGGAATGTACAAACCCCACTGCGGAATCGATAATTTGCTGATGTCCTGGAGCCACGATGAGTATCTGTATCAATTCCTGCTTCATAACAAGTCTACGATACCTGAGAAGGGTCTATACATGATCAGGTACCACTCGTTCTACCCTTGGCACGCAGGCGGAGACTACCGCCATCTGAGCAATGAAAAAGACGAAGAAATTCTAAAATGGGTGACCGAATTTAACAAATATGATTTGTACACGAAGAGTGAGAAGGTGCCGGACATTGAGGCCCTTTGGCCGTACTACGAAAAGCTCATTGAGAAGTACATCCCCGGAATCTGCGAATGGTGA
Protein
MAAEVEKKPVSMIDPSQLLRPEPTFNDKPIDAFRDYSVDDTDPIKERVRRTYYLMHSNVTVDLVKQKREKWLKFNHFKATVKDALIKLNELVDESDPDTDLPNIVHAFQTAERIREDHPDEDWFHLTGLIHDLGKVMAFYDEPQWCVVGDTFPVGCKWADSIVYGPESFKDNPDTYNPKYNTKYGMYKPHCGIDNLLMSWSHDEYLYQFLLHNKSTIPEKGLYMIRYHSFYPWHAGGDYRHLSNEKDEEILKWVTEFNKYDLYTKSEKVPDIEALWPYYEKLIEKYIPGICEW
Summary
Uniprot
Q1HPR7
H9J8E4
A0A2H1W206
A0A212FMQ6
A0A2A4JQ24
A0A2W1BFI2
+ More
A0A437BFA4 A0A2W1BGE4 A0A3S2NEM0 A0A2H1W0B3 A0A2A4JQH2 A0A0N1I6Y1 A0A0T6AUM3 A0A1W4WZ62 A0A1B6L1W8 A0A1B6LQ55 A0A1B6MT11 K7IS44 D2A5T3 A0A0L0BXQ2 A0A1B0D973 A0A1B6BYJ6 V5GZ83 A0A2M4AG82 A0A2M4CTA5 A0A2M4CTC9 A0A2M4AG99 E2BWJ0 A0A2M4BVN6 A0A2M4BWL7 A0A2J7QAN1 Q16XZ8 A0A2M4AIE5 T1PBA6 A0A1Q3FWF3 A0A212FMS5 A0A067QRT6 A0A1S4FK57 E2AQZ4 A0A2M3Z3D7 A0A1L8E970 A0A195EL30 A0A088AUW4 A0A026W1N0 A0A023ENT5 A0A194Q195 A0A2M3Z4D5 A0A182N6D3 A0A023ENU1 A0A182G8I1 Q7PQI1 A0A182QLX0 A0A158P092 A0A182I9A0 A0A195CIC2 A0A182WVM1 A0A182UXW4 A0A182KXQ7 A0A1Y1LG78 A0A195BMT6 A0A151WRF1 V9IKP5 A0A182TPP6 A0A182MS79 A0A1I8P389 A0A182GAG9 W8B1U9 A0A1B0BII8 A0A1L8DCQ1 A0A1B0A1U5 N6URW2 A0A1A9YM72 D3TLA8 A0A1A9W1Z8 A0A1A9VSY6 B4L3T3 A0A034VWK7 B4GR69 A0A0K8U4P5 B4MD17 A0A182F813 B5DQN8 A0A0J7KPZ7 B3M7V4 B4N3S8 A0A1J1IUF8 A0A0M4EHM5 A0A182IZS4 A0A1W4UWC5 B4PGA5 Q9VTU9 A0A226EW93 A0A232EW30 B3NH78 B4HFJ9 A0A2A3E1F6 A0A182RBG2 A0A182YIC0 E0W0V9 B4QR69
A0A437BFA4 A0A2W1BGE4 A0A3S2NEM0 A0A2H1W0B3 A0A2A4JQH2 A0A0N1I6Y1 A0A0T6AUM3 A0A1W4WZ62 A0A1B6L1W8 A0A1B6LQ55 A0A1B6MT11 K7IS44 D2A5T3 A0A0L0BXQ2 A0A1B0D973 A0A1B6BYJ6 V5GZ83 A0A2M4AG82 A0A2M4CTA5 A0A2M4CTC9 A0A2M4AG99 E2BWJ0 A0A2M4BVN6 A0A2M4BWL7 A0A2J7QAN1 Q16XZ8 A0A2M4AIE5 T1PBA6 A0A1Q3FWF3 A0A212FMS5 A0A067QRT6 A0A1S4FK57 E2AQZ4 A0A2M3Z3D7 A0A1L8E970 A0A195EL30 A0A088AUW4 A0A026W1N0 A0A023ENT5 A0A194Q195 A0A2M3Z4D5 A0A182N6D3 A0A023ENU1 A0A182G8I1 Q7PQI1 A0A182QLX0 A0A158P092 A0A182I9A0 A0A195CIC2 A0A182WVM1 A0A182UXW4 A0A182KXQ7 A0A1Y1LG78 A0A195BMT6 A0A151WRF1 V9IKP5 A0A182TPP6 A0A182MS79 A0A1I8P389 A0A182GAG9 W8B1U9 A0A1B0BII8 A0A1L8DCQ1 A0A1B0A1U5 N6URW2 A0A1A9YM72 D3TLA8 A0A1A9W1Z8 A0A1A9VSY6 B4L3T3 A0A034VWK7 B4GR69 A0A0K8U4P5 B4MD17 A0A182F813 B5DQN8 A0A0J7KPZ7 B3M7V4 B4N3S8 A0A1J1IUF8 A0A0M4EHM5 A0A182IZS4 A0A1W4UWC5 B4PGA5 Q9VTU9 A0A226EW93 A0A232EW30 B3NH78 B4HFJ9 A0A2A3E1F6 A0A182RBG2 A0A182YIC0 E0W0V9 B4QR69
Pubmed
19121390
22118469
28756777
26354079
20075255
18362917
+ More
19820115 26108605 20798317 17510324 25315136 24845553 24508170 30249741 24945155 26483478 12364791 14747013 17210077 21347285 20966253 28004739 24495485 23537049 20353571 17994087 25348373 15632085 18057021 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 25244985 20566863 22936249
19820115 26108605 20798317 17510324 25315136 24845553 24508170 30249741 24945155 26483478 12364791 14747013 17210077 21347285 20966253 28004739 24495485 23537049 20353571 17994087 25348373 15632085 18057021 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 25244985 20566863 22936249
EMBL
DQ443335
ABF51424.1
BABH01027310
BABH01027311
BABH01027312
BABH01027313
+ More
ODYU01005569 SOQ46544.1 AGBW02007651 OWR55042.1 NWSH01000886 PCG73694.1 KZ150081 PZC73869.1 RSAL01000071 RVE49116.1 PZC73868.1 RVE49115.1 SOQ46545.1 PCG73693.1 KQ460651 KPJ12989.1 LJIG01022771 KRT78802.1 GEBQ01022278 JAT17699.1 GEBQ01014218 JAT25759.1 GEBQ01000906 JAT39071.1 KQ971345 EFA05415.1 JRES01001169 KNC24828.1 AJVK01004263 AJVK01004264 GEDC01030970 GEDC01004203 JAS06328.1 JAS33095.1 GALX01002668 JAB65798.1 GGFK01006465 MBW39786.1 GGFL01004404 MBW68582.1 GGFL01004405 MBW68583.1 GGFK01006478 MBW39799.1 GL451131 EFN79946.1 GGFJ01007978 MBW57119.1 GGFJ01007977 MBW57118.1 NEVH01016329 PNF25634.1 CH477528 EAT39505.1 GGFK01007181 MBW40502.1 KA646012 AFP60641.1 GFDL01003160 JAV31885.1 OWR55041.1 KK853845 KDQ71560.1 GL441846 EFN64140.1 GGFM01002283 MBW23034.1 GFDG01003667 JAV15132.1 KQ978747 KYN28647.1 KK107488 QOIP01000012 EZA49933.1 RLU15740.1 GAPW01002992 JAC10606.1 KQ459582 KPI99093.1 GGFM01002629 MBW23380.1 GAPW01002993 JAC10605.1 JXUM01047801 KQ561533 KXJ78246.1 AAAB01008888 EAA08814.4 AXCN02000284 ADTU01004642 APCN01003239 KQ977720 KYN00473.1 GEZM01058133 JAV71881.1 KQ976433 KYM87290.1 KQ982807 KYQ50381.1 JR049182 AEY60889.1 AXCM01001551 JXUM01050913 KQ561667 KXJ77859.1 GAMC01014113 JAB92442.1 JXJN01014969 GFDF01009841 JAV04243.1 APGK01019139 KB740092 KB632429 ENN81472.1 ERL95421.1 CCAG010011658 EZ422210 ADD18486.1 CH933810 EDW07211.1 GAKP01013024 JAC45928.1 CH479188 EDW40254.1 GDHF01030786 JAI21528.1 CH940660 EDW58089.1 CH379069 EDY73461.1 LBMM01004500 KMQ92351.1 CH902618 EDV38827.1 CH964095 EDW79283.1 EDW79285.1 CVRI01000060 CRL03853.1 CP012528 ALC48166.1 CM000159 EDW94269.1 AE014296 BT011521 AAF49947.3 AAS15657.1 LNIX01000002 OXA61111.1 NNAY01001922 OXU22558.1 CH954178 EDV51535.1 CH480815 EDW41230.1 KZ288455 PBC25527.1 DS235862 EEB19265.1 CM000363 CM002912 EDX10199.1 KMY99181.1
ODYU01005569 SOQ46544.1 AGBW02007651 OWR55042.1 NWSH01000886 PCG73694.1 KZ150081 PZC73869.1 RSAL01000071 RVE49116.1 PZC73868.1 RVE49115.1 SOQ46545.1 PCG73693.1 KQ460651 KPJ12989.1 LJIG01022771 KRT78802.1 GEBQ01022278 JAT17699.1 GEBQ01014218 JAT25759.1 GEBQ01000906 JAT39071.1 KQ971345 EFA05415.1 JRES01001169 KNC24828.1 AJVK01004263 AJVK01004264 GEDC01030970 GEDC01004203 JAS06328.1 JAS33095.1 GALX01002668 JAB65798.1 GGFK01006465 MBW39786.1 GGFL01004404 MBW68582.1 GGFL01004405 MBW68583.1 GGFK01006478 MBW39799.1 GL451131 EFN79946.1 GGFJ01007978 MBW57119.1 GGFJ01007977 MBW57118.1 NEVH01016329 PNF25634.1 CH477528 EAT39505.1 GGFK01007181 MBW40502.1 KA646012 AFP60641.1 GFDL01003160 JAV31885.1 OWR55041.1 KK853845 KDQ71560.1 GL441846 EFN64140.1 GGFM01002283 MBW23034.1 GFDG01003667 JAV15132.1 KQ978747 KYN28647.1 KK107488 QOIP01000012 EZA49933.1 RLU15740.1 GAPW01002992 JAC10606.1 KQ459582 KPI99093.1 GGFM01002629 MBW23380.1 GAPW01002993 JAC10605.1 JXUM01047801 KQ561533 KXJ78246.1 AAAB01008888 EAA08814.4 AXCN02000284 ADTU01004642 APCN01003239 KQ977720 KYN00473.1 GEZM01058133 JAV71881.1 KQ976433 KYM87290.1 KQ982807 KYQ50381.1 JR049182 AEY60889.1 AXCM01001551 JXUM01050913 KQ561667 KXJ77859.1 GAMC01014113 JAB92442.1 JXJN01014969 GFDF01009841 JAV04243.1 APGK01019139 KB740092 KB632429 ENN81472.1 ERL95421.1 CCAG010011658 EZ422210 ADD18486.1 CH933810 EDW07211.1 GAKP01013024 JAC45928.1 CH479188 EDW40254.1 GDHF01030786 JAI21528.1 CH940660 EDW58089.1 CH379069 EDY73461.1 LBMM01004500 KMQ92351.1 CH902618 EDV38827.1 CH964095 EDW79283.1 EDW79285.1 CVRI01000060 CRL03853.1 CP012528 ALC48166.1 CM000159 EDW94269.1 AE014296 BT011521 AAF49947.3 AAS15657.1 LNIX01000002 OXA61111.1 NNAY01001922 OXU22558.1 CH954178 EDV51535.1 CH480815 EDW41230.1 KZ288455 PBC25527.1 DS235862 EEB19265.1 CM000363 CM002912 EDX10199.1 KMY99181.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000283053
UP000053240
UP000192223
+ More
UP000002358 UP000007266 UP000037069 UP000092462 UP000008237 UP000235965 UP000008820 UP000095301 UP000027135 UP000000311 UP000078492 UP000005203 UP000053097 UP000279307 UP000053268 UP000075884 UP000069940 UP000249989 UP000007062 UP000075886 UP000005205 UP000075840 UP000078542 UP000076407 UP000075903 UP000075882 UP000078540 UP000075809 UP000075902 UP000075883 UP000095300 UP000092460 UP000092445 UP000019118 UP000030742 UP000092443 UP000092444 UP000091820 UP000078200 UP000009192 UP000008744 UP000008792 UP000069272 UP000001819 UP000036403 UP000007801 UP000007798 UP000183832 UP000092553 UP000075880 UP000192221 UP000002282 UP000000803 UP000198287 UP000215335 UP000008711 UP000001292 UP000242457 UP000075900 UP000076408 UP000009046 UP000000304
UP000002358 UP000007266 UP000037069 UP000092462 UP000008237 UP000235965 UP000008820 UP000095301 UP000027135 UP000000311 UP000078492 UP000005203 UP000053097 UP000279307 UP000053268 UP000075884 UP000069940 UP000249989 UP000007062 UP000075886 UP000005205 UP000075840 UP000078542 UP000076407 UP000075903 UP000075882 UP000078540 UP000075809 UP000075902 UP000075883 UP000095300 UP000092460 UP000092445 UP000019118 UP000030742 UP000092443 UP000092444 UP000091820 UP000078200 UP000009192 UP000008744 UP000008792 UP000069272 UP000001819 UP000036403 UP000007801 UP000007798 UP000183832 UP000092553 UP000075880 UP000192221 UP000002282 UP000000803 UP000198287 UP000215335 UP000008711 UP000001292 UP000242457 UP000075900 UP000076408 UP000009046 UP000000304
PRIDE
Interpro
Gene 3D
ProteinModelPortal
Q1HPR7
H9J8E4
A0A2H1W206
A0A212FMQ6
A0A2A4JQ24
A0A2W1BFI2
+ More
A0A437BFA4 A0A2W1BGE4 A0A3S2NEM0 A0A2H1W0B3 A0A2A4JQH2 A0A0N1I6Y1 A0A0T6AUM3 A0A1W4WZ62 A0A1B6L1W8 A0A1B6LQ55 A0A1B6MT11 K7IS44 D2A5T3 A0A0L0BXQ2 A0A1B0D973 A0A1B6BYJ6 V5GZ83 A0A2M4AG82 A0A2M4CTA5 A0A2M4CTC9 A0A2M4AG99 E2BWJ0 A0A2M4BVN6 A0A2M4BWL7 A0A2J7QAN1 Q16XZ8 A0A2M4AIE5 T1PBA6 A0A1Q3FWF3 A0A212FMS5 A0A067QRT6 A0A1S4FK57 E2AQZ4 A0A2M3Z3D7 A0A1L8E970 A0A195EL30 A0A088AUW4 A0A026W1N0 A0A023ENT5 A0A194Q195 A0A2M3Z4D5 A0A182N6D3 A0A023ENU1 A0A182G8I1 Q7PQI1 A0A182QLX0 A0A158P092 A0A182I9A0 A0A195CIC2 A0A182WVM1 A0A182UXW4 A0A182KXQ7 A0A1Y1LG78 A0A195BMT6 A0A151WRF1 V9IKP5 A0A182TPP6 A0A182MS79 A0A1I8P389 A0A182GAG9 W8B1U9 A0A1B0BII8 A0A1L8DCQ1 A0A1B0A1U5 N6URW2 A0A1A9YM72 D3TLA8 A0A1A9W1Z8 A0A1A9VSY6 B4L3T3 A0A034VWK7 B4GR69 A0A0K8U4P5 B4MD17 A0A182F813 B5DQN8 A0A0J7KPZ7 B3M7V4 B4N3S8 A0A1J1IUF8 A0A0M4EHM5 A0A182IZS4 A0A1W4UWC5 B4PGA5 Q9VTU9 A0A226EW93 A0A232EW30 B3NH78 B4HFJ9 A0A2A3E1F6 A0A182RBG2 A0A182YIC0 E0W0V9 B4QR69
A0A437BFA4 A0A2W1BGE4 A0A3S2NEM0 A0A2H1W0B3 A0A2A4JQH2 A0A0N1I6Y1 A0A0T6AUM3 A0A1W4WZ62 A0A1B6L1W8 A0A1B6LQ55 A0A1B6MT11 K7IS44 D2A5T3 A0A0L0BXQ2 A0A1B0D973 A0A1B6BYJ6 V5GZ83 A0A2M4AG82 A0A2M4CTA5 A0A2M4CTC9 A0A2M4AG99 E2BWJ0 A0A2M4BVN6 A0A2M4BWL7 A0A2J7QAN1 Q16XZ8 A0A2M4AIE5 T1PBA6 A0A1Q3FWF3 A0A212FMS5 A0A067QRT6 A0A1S4FK57 E2AQZ4 A0A2M3Z3D7 A0A1L8E970 A0A195EL30 A0A088AUW4 A0A026W1N0 A0A023ENT5 A0A194Q195 A0A2M3Z4D5 A0A182N6D3 A0A023ENU1 A0A182G8I1 Q7PQI1 A0A182QLX0 A0A158P092 A0A182I9A0 A0A195CIC2 A0A182WVM1 A0A182UXW4 A0A182KXQ7 A0A1Y1LG78 A0A195BMT6 A0A151WRF1 V9IKP5 A0A182TPP6 A0A182MS79 A0A1I8P389 A0A182GAG9 W8B1U9 A0A1B0BII8 A0A1L8DCQ1 A0A1B0A1U5 N6URW2 A0A1A9YM72 D3TLA8 A0A1A9W1Z8 A0A1A9VSY6 B4L3T3 A0A034VWK7 B4GR69 A0A0K8U4P5 B4MD17 A0A182F813 B5DQN8 A0A0J7KPZ7 B3M7V4 B4N3S8 A0A1J1IUF8 A0A0M4EHM5 A0A182IZS4 A0A1W4UWC5 B4PGA5 Q9VTU9 A0A226EW93 A0A232EW30 B3NH78 B4HFJ9 A0A2A3E1F6 A0A182RBG2 A0A182YIC0 E0W0V9 B4QR69
PDB
3BXD
E-value=5.45939e-91,
Score=851
Ontologies
PATHWAY
GO
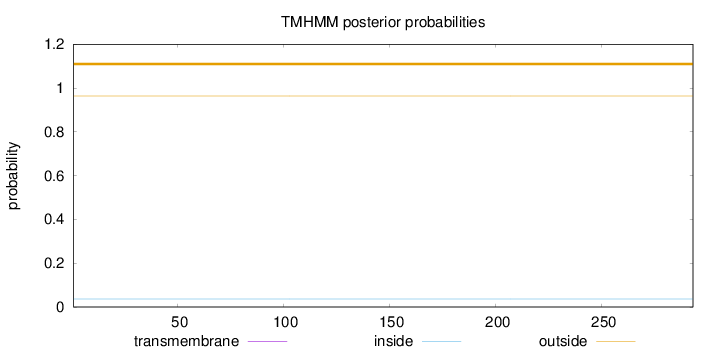
Topology
Length:
293
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03663
outside
1 - 293
Population Genetic Test Statistics
Pi
239.54873
Theta
149.906218
Tajima's D
3.690593
CLR
0.075327
CSRT
0.99630018499075
Interpretation
Uncertain
