Pre Gene Modal
BGIBMGA013044
Annotation
p270_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.532
Sequence
CDS
ATGGCTCCAACAACAGTGGTCGAAGACAGATTGACGTCTGGACACCGGCTGTCGCACCCCCCGTCTGGAGATGAGGTATACGTCACCGGAGTCTCGGGGTACTTCCCGGATTCCGATTCAGTGAAACATCTACAAGAAAACCTTTTTAATAAGGTGGATTTAATCTCCTCGGATGACAGACGTTGGAAACTGGCACACCCTGAGATACCGCAGCGGACCGGCAAGATCAACAATGTCAACAAGTTCGACGCCTCCTTCTTCGGCGTCCACTACAAGCAGGCGCACACCATGGACCCCATGTGCAGGATCTTATTGGAGAAGGCCTATGAAGCTATCGTTGATGCCGGCCTAAACCCAAAAGAGCTTCGTGACACGAAAACGGGTGTTTTCATTGGCGCTTGCTTCTCCGAATCCGAGAAAACTTGGTTCTACGAAAAGATGCAGGTTAATGGATTCGGTATCACTGGGTAA
Protein
MAPTTVVEDRLTSGHRLSHPPSGDEVYVTGVSGYFPDSDSVKHLQENLFNKVDLISSDDRRWKLAHPEIPQRTGKINNVNKFDASFFGVHYKQAHTMDPMCRILLEKAYEAIVDAGLNPKELRDTKTGVFIGACFSESEKTWFYEKMQVNGFGITG
Summary
Similarity
Belongs to the thiolase-like superfamily. Beta-ketoacyl-ACP synthases family.
Uniprot
O01678
A0A2W1BEL1
A0A3S2LA71
A0A2A4J2W6
A0A2A4J169
A0A212FMU5
+ More
A0A194Q6Q7 A0A0N1IP09 A0A2H1WS03 A0A2H1VF66 A0A2H1VGF0 H9JU32 A0A2H1VFB6 A0A1S4EB00 A0A1B6GA85 A0A1B6IHH2 A0A1B6JP24 A0A151X2S2 A0A1B6CGM1 A0A2S2RA89 A0A2P8XKN7 A0A0C9Q3R4 A0A0J7NXT7 A0A2H8TWM7 A0A232EXC2 K7IZC9 E0VU95 A0A195CP13 A0A195F360 A0A151JRB8 J9K3D3 A0A0G3YL52 K7IZC8 F4WIF1 A0A232F4D3 A0A158NZW8 A0A195BMY6 A0A067RHL3 E2ADN8 A0A2R7W720 E2B583 A0A026WNV9 A0A3L8DXR9 A0A1W4XV87 E9IT55 A0A0A9Y5L4 A0A146M080 A0A1B0CP42 B0WMC9 A0A182GNP7 A0A1Q3FS82 A0A1S4FIT0 Q16ZI9 A0A182N5X9 A0A084VRX8 A0A182QDS2 A0A182J9B8 A0A182FQK1 A0A182KM38 A0A182TPE9 Q7PYE4 A0A182I0F0 A0A182V5F6 A0A182JQR8 A0A182XBL3 A0A182R1J1 A0A182YJT7 A0A310SRI2 A0A0L7R328 A0A0M8ZRC2 A0A087ZKN3 W5J299 A0A1J1IGX7 A0A1A9XHP8 A0A182PC27 A0A182VQY2 A0A182M3X3 A0A154PMH1 A0A2J7PQX9 A0A2A3E4Z2 A0A2H1WVG7 A0A088ALX8 B4LXF7 A0A034VKR9 A0A034VP61 A0A1B0D3T5 W8ARH4 A0A0A1XSP3 A0A0K8UG84 A0A0K8U8C9 A0A336M5W9 A0A0M4EN44 E2BR61 A0A1B0G1J3 A0A1B0AA77 B4KAW7 A0A0L0BQ63 E2BR60 B4NIW4
A0A194Q6Q7 A0A0N1IP09 A0A2H1WS03 A0A2H1VF66 A0A2H1VGF0 H9JU32 A0A2H1VFB6 A0A1S4EB00 A0A1B6GA85 A0A1B6IHH2 A0A1B6JP24 A0A151X2S2 A0A1B6CGM1 A0A2S2RA89 A0A2P8XKN7 A0A0C9Q3R4 A0A0J7NXT7 A0A2H8TWM7 A0A232EXC2 K7IZC9 E0VU95 A0A195CP13 A0A195F360 A0A151JRB8 J9K3D3 A0A0G3YL52 K7IZC8 F4WIF1 A0A232F4D3 A0A158NZW8 A0A195BMY6 A0A067RHL3 E2ADN8 A0A2R7W720 E2B583 A0A026WNV9 A0A3L8DXR9 A0A1W4XV87 E9IT55 A0A0A9Y5L4 A0A146M080 A0A1B0CP42 B0WMC9 A0A182GNP7 A0A1Q3FS82 A0A1S4FIT0 Q16ZI9 A0A182N5X9 A0A084VRX8 A0A182QDS2 A0A182J9B8 A0A182FQK1 A0A182KM38 A0A182TPE9 Q7PYE4 A0A182I0F0 A0A182V5F6 A0A182JQR8 A0A182XBL3 A0A182R1J1 A0A182YJT7 A0A310SRI2 A0A0L7R328 A0A0M8ZRC2 A0A087ZKN3 W5J299 A0A1J1IGX7 A0A1A9XHP8 A0A182PC27 A0A182VQY2 A0A182M3X3 A0A154PMH1 A0A2J7PQX9 A0A2A3E4Z2 A0A2H1WVG7 A0A088ALX8 B4LXF7 A0A034VKR9 A0A034VP61 A0A1B0D3T5 W8ARH4 A0A0A1XSP3 A0A0K8UG84 A0A0K8U8C9 A0A336M5W9 A0A0M4EN44 E2BR61 A0A1B0G1J3 A0A1B0AA77 B4KAW7 A0A0L0BQ63 E2BR60 B4NIW4
Pubmed
EMBL
U67867
AAB53258.1
KZ150116
PZC73268.1
RSAL01000071
RVE49124.1
+ More
NWSH01003873 PCG65733.1 PCG65731.1 AGBW02007651 OWR55047.1 KQ459582 KPI99075.1 KQ460651 KPJ13003.1 ODYU01010623 SOQ55855.1 ODYU01002244 SOQ39469.1 ODYU01002246 SOQ39472.1 BABH01003984 BABH01003985 ODYU01002245 SOQ39471.1 GECZ01010430 JAS59339.1 GECU01021338 JAS86368.1 GECU01006695 JAT01012.1 KQ982567 KYQ54716.1 GEDC01024813 JAS12485.1 GGMS01017661 MBY86864.1 PYGN01001841 PSN32561.1 GBYB01008708 JAG78475.1 LBMM01000911 KMQ97230.1 GFXV01006890 MBW18695.1 NNAY01001760 OXU23005.1 DS235784 EEB16951.1 KQ977574 KYN01849.1 KQ981856 KYN34617.1 KQ978607 KYN29892.1 ABLF02036897 KM507100 AKM28424.1 GL888175 EGI66082.1 NNAY01001038 OXU25389.1 ADTU01005072 ADTU01005073 ADTU01005074 ADTU01005075 KQ976440 KYM86527.1 KK852680 KDR18643.1 GL438820 EFN68400.1 KK854269 PTY14005.1 GL445739 EFN89136.1 KK107139 EZA57700.1 QOIP01000003 RLU24885.1 GL765434 EFZ16301.1 GBHO01016150 GBRD01007997 JAG27454.1 JAG57824.1 GDHC01005385 JAQ13244.1 AJWK01021412 DS231997 EDS30986.1 JXUM01076840 KQ562972 KXJ74732.1 GFDL01004709 JAV30336.1 CH477489 EAT40085.1 ATLV01015783 ATLV01015784 KE525036 KFB40722.1 AXCN02000191 AAAB01008987 EAA01044.4 APCN01002072 KQ761246 OAD57941.1 KQ414663 KOC65238.1 KQ435885 KOX69550.1 ADMH02002134 ETN58382.1 CVRI01000051 CRK99509.1 AXCM01001692 KQ434960 KZC12654.1 NEVH01022635 PNF18739.1 KZ288370 PBC26760.1 ODYU01011362 SOQ57060.1 CH940650 EDW67835.2 GAKP01015046 JAC43906.1 GAKP01015045 JAC43907.1 AJVK01011044 GAMC01019372 JAB87183.1 GBXI01000719 JAD13573.1 GDHF01026736 JAI25578.1 GDHF01029400 JAI22914.1 UFQT01000602 SSX25625.1 CP012526 ALC46026.1 GL449898 EFN81867.1 CCAG010017947 CH933806 EDW14645.2 JRES01001625 KNC21349.1 EFN81866.1 CH964272 EDW84866.1
NWSH01003873 PCG65733.1 PCG65731.1 AGBW02007651 OWR55047.1 KQ459582 KPI99075.1 KQ460651 KPJ13003.1 ODYU01010623 SOQ55855.1 ODYU01002244 SOQ39469.1 ODYU01002246 SOQ39472.1 BABH01003984 BABH01003985 ODYU01002245 SOQ39471.1 GECZ01010430 JAS59339.1 GECU01021338 JAS86368.1 GECU01006695 JAT01012.1 KQ982567 KYQ54716.1 GEDC01024813 JAS12485.1 GGMS01017661 MBY86864.1 PYGN01001841 PSN32561.1 GBYB01008708 JAG78475.1 LBMM01000911 KMQ97230.1 GFXV01006890 MBW18695.1 NNAY01001760 OXU23005.1 DS235784 EEB16951.1 KQ977574 KYN01849.1 KQ981856 KYN34617.1 KQ978607 KYN29892.1 ABLF02036897 KM507100 AKM28424.1 GL888175 EGI66082.1 NNAY01001038 OXU25389.1 ADTU01005072 ADTU01005073 ADTU01005074 ADTU01005075 KQ976440 KYM86527.1 KK852680 KDR18643.1 GL438820 EFN68400.1 KK854269 PTY14005.1 GL445739 EFN89136.1 KK107139 EZA57700.1 QOIP01000003 RLU24885.1 GL765434 EFZ16301.1 GBHO01016150 GBRD01007997 JAG27454.1 JAG57824.1 GDHC01005385 JAQ13244.1 AJWK01021412 DS231997 EDS30986.1 JXUM01076840 KQ562972 KXJ74732.1 GFDL01004709 JAV30336.1 CH477489 EAT40085.1 ATLV01015783 ATLV01015784 KE525036 KFB40722.1 AXCN02000191 AAAB01008987 EAA01044.4 APCN01002072 KQ761246 OAD57941.1 KQ414663 KOC65238.1 KQ435885 KOX69550.1 ADMH02002134 ETN58382.1 CVRI01000051 CRK99509.1 AXCM01001692 KQ434960 KZC12654.1 NEVH01022635 PNF18739.1 KZ288370 PBC26760.1 ODYU01011362 SOQ57060.1 CH940650 EDW67835.2 GAKP01015046 JAC43906.1 GAKP01015045 JAC43907.1 AJVK01011044 GAMC01019372 JAB87183.1 GBXI01000719 JAD13573.1 GDHF01026736 JAI25578.1 GDHF01029400 JAI22914.1 UFQT01000602 SSX25625.1 CP012526 ALC46026.1 GL449898 EFN81867.1 CCAG010017947 CH933806 EDW14645.2 JRES01001625 KNC21349.1 EFN81866.1 CH964272 EDW84866.1
Proteomes
UP000283053
UP000218220
UP000007151
UP000053268
UP000053240
UP000005204
+ More
UP000079169 UP000075809 UP000245037 UP000036403 UP000215335 UP000002358 UP000009046 UP000078542 UP000078541 UP000078492 UP000007819 UP000007755 UP000005205 UP000078540 UP000027135 UP000000311 UP000008237 UP000053097 UP000279307 UP000192223 UP000092461 UP000002320 UP000069940 UP000249989 UP000008820 UP000075884 UP000030765 UP000075886 UP000075880 UP000069272 UP000075882 UP000075902 UP000007062 UP000075840 UP000075903 UP000075881 UP000076407 UP000075900 UP000076408 UP000053825 UP000053105 UP000000673 UP000183832 UP000092443 UP000075885 UP000075920 UP000075883 UP000076502 UP000235965 UP000242457 UP000005203 UP000008792 UP000092462 UP000092553 UP000092444 UP000092445 UP000009192 UP000037069 UP000007798
UP000079169 UP000075809 UP000245037 UP000036403 UP000215335 UP000002358 UP000009046 UP000078542 UP000078541 UP000078492 UP000007819 UP000007755 UP000005205 UP000078540 UP000027135 UP000000311 UP000008237 UP000053097 UP000279307 UP000192223 UP000092461 UP000002320 UP000069940 UP000249989 UP000008820 UP000075884 UP000030765 UP000075886 UP000075880 UP000069272 UP000075882 UP000075902 UP000007062 UP000075840 UP000075903 UP000075881 UP000076407 UP000075900 UP000076408 UP000053825 UP000053105 UP000000673 UP000183832 UP000092443 UP000075885 UP000075920 UP000075883 UP000076502 UP000235965 UP000242457 UP000005203 UP000008792 UP000092462 UP000092553 UP000092444 UP000092445 UP000009192 UP000037069 UP000007798
PRIDE
Pfam
Interpro
IPR013968
PKS_KR
+ More
IPR014030 Ketoacyl_synth_N
IPR020843 PKS_ER
IPR016039 Thiolase-like
IPR020801 PKS_acyl_transferase
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR029058 AB_hydrolase
IPR020807 PKS_dehydratase
IPR001227 Ac_transferase_dom_sf
IPR016036 Malonyl_transacylase_ACP-bd
IPR036736 ACP-like_sf
IPR016035 Acyl_Trfase/lysoPLipase
IPR014031 Ketoacyl_synth_C
IPR023102 Fatty_acid_synthase_dom_2
IPR009081 PP-bd_ACP
IPR042104 PKS_dehydratase_sf
IPR020806 PKS_PP-bd
IPR011032 GroES-like_sf
IPR032821 KAsynt_C_assoc
IPR013149 ADH_C
IPR001031 Thioesterase
IPR014043 Acyl_transferase
IPR036291 NAD(P)-bd_dom_sf
IPR018201 Ketoacyl_synth_AS
IPR000999 RNase_III_dom
IPR036389 RNase_III_sf
IPR006162 Ppantetheine_attach_site
IPR014030 Ketoacyl_synth_N
IPR020843 PKS_ER
IPR016039 Thiolase-like
IPR020801 PKS_acyl_transferase
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR029058 AB_hydrolase
IPR020807 PKS_dehydratase
IPR001227 Ac_transferase_dom_sf
IPR016036 Malonyl_transacylase_ACP-bd
IPR036736 ACP-like_sf
IPR016035 Acyl_Trfase/lysoPLipase
IPR014031 Ketoacyl_synth_C
IPR023102 Fatty_acid_synthase_dom_2
IPR009081 PP-bd_ACP
IPR042104 PKS_dehydratase_sf
IPR020806 PKS_PP-bd
IPR011032 GroES-like_sf
IPR032821 KAsynt_C_assoc
IPR013149 ADH_C
IPR001031 Thioesterase
IPR014043 Acyl_transferase
IPR036291 NAD(P)-bd_dom_sf
IPR018201 Ketoacyl_synth_AS
IPR000999 RNase_III_dom
IPR036389 RNase_III_sf
IPR006162 Ppantetheine_attach_site
SUPFAM
CDD
ProteinModelPortal
O01678
A0A2W1BEL1
A0A3S2LA71
A0A2A4J2W6
A0A2A4J169
A0A212FMU5
+ More
A0A194Q6Q7 A0A0N1IP09 A0A2H1WS03 A0A2H1VF66 A0A2H1VGF0 H9JU32 A0A2H1VFB6 A0A1S4EB00 A0A1B6GA85 A0A1B6IHH2 A0A1B6JP24 A0A151X2S2 A0A1B6CGM1 A0A2S2RA89 A0A2P8XKN7 A0A0C9Q3R4 A0A0J7NXT7 A0A2H8TWM7 A0A232EXC2 K7IZC9 E0VU95 A0A195CP13 A0A195F360 A0A151JRB8 J9K3D3 A0A0G3YL52 K7IZC8 F4WIF1 A0A232F4D3 A0A158NZW8 A0A195BMY6 A0A067RHL3 E2ADN8 A0A2R7W720 E2B583 A0A026WNV9 A0A3L8DXR9 A0A1W4XV87 E9IT55 A0A0A9Y5L4 A0A146M080 A0A1B0CP42 B0WMC9 A0A182GNP7 A0A1Q3FS82 A0A1S4FIT0 Q16ZI9 A0A182N5X9 A0A084VRX8 A0A182QDS2 A0A182J9B8 A0A182FQK1 A0A182KM38 A0A182TPE9 Q7PYE4 A0A182I0F0 A0A182V5F6 A0A182JQR8 A0A182XBL3 A0A182R1J1 A0A182YJT7 A0A310SRI2 A0A0L7R328 A0A0M8ZRC2 A0A087ZKN3 W5J299 A0A1J1IGX7 A0A1A9XHP8 A0A182PC27 A0A182VQY2 A0A182M3X3 A0A154PMH1 A0A2J7PQX9 A0A2A3E4Z2 A0A2H1WVG7 A0A088ALX8 B4LXF7 A0A034VKR9 A0A034VP61 A0A1B0D3T5 W8ARH4 A0A0A1XSP3 A0A0K8UG84 A0A0K8U8C9 A0A336M5W9 A0A0M4EN44 E2BR61 A0A1B0G1J3 A0A1B0AA77 B4KAW7 A0A0L0BQ63 E2BR60 B4NIW4
A0A194Q6Q7 A0A0N1IP09 A0A2H1WS03 A0A2H1VF66 A0A2H1VGF0 H9JU32 A0A2H1VFB6 A0A1S4EB00 A0A1B6GA85 A0A1B6IHH2 A0A1B6JP24 A0A151X2S2 A0A1B6CGM1 A0A2S2RA89 A0A2P8XKN7 A0A0C9Q3R4 A0A0J7NXT7 A0A2H8TWM7 A0A232EXC2 K7IZC9 E0VU95 A0A195CP13 A0A195F360 A0A151JRB8 J9K3D3 A0A0G3YL52 K7IZC8 F4WIF1 A0A232F4D3 A0A158NZW8 A0A195BMY6 A0A067RHL3 E2ADN8 A0A2R7W720 E2B583 A0A026WNV9 A0A3L8DXR9 A0A1W4XV87 E9IT55 A0A0A9Y5L4 A0A146M080 A0A1B0CP42 B0WMC9 A0A182GNP7 A0A1Q3FS82 A0A1S4FIT0 Q16ZI9 A0A182N5X9 A0A084VRX8 A0A182QDS2 A0A182J9B8 A0A182FQK1 A0A182KM38 A0A182TPE9 Q7PYE4 A0A182I0F0 A0A182V5F6 A0A182JQR8 A0A182XBL3 A0A182R1J1 A0A182YJT7 A0A310SRI2 A0A0L7R328 A0A0M8ZRC2 A0A087ZKN3 W5J299 A0A1J1IGX7 A0A1A9XHP8 A0A182PC27 A0A182VQY2 A0A182M3X3 A0A154PMH1 A0A2J7PQX9 A0A2A3E4Z2 A0A2H1WVG7 A0A088ALX8 B4LXF7 A0A034VKR9 A0A034VP61 A0A1B0D3T5 W8ARH4 A0A0A1XSP3 A0A0K8UG84 A0A0K8U8C9 A0A336M5W9 A0A0M4EN44 E2BR61 A0A1B0G1J3 A0A1B0AA77 B4KAW7 A0A0L0BQ63 E2BR60 B4NIW4
PDB
3HHD
E-value=1.28584e-34,
Score=361
Ontologies
PATHWAY
GO
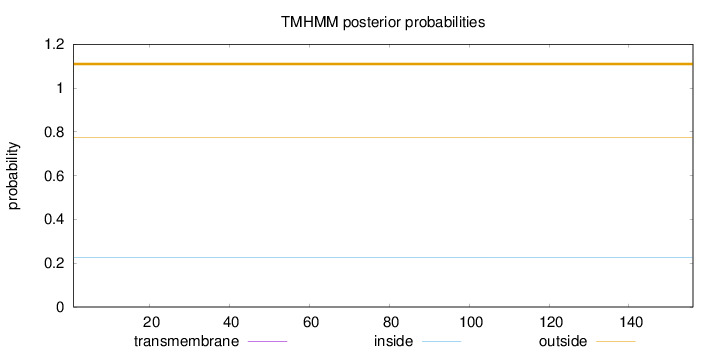
Topology
Length:
156
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00036
Exp number, first 60 AAs:
0
Total prob of N-in:
0.22611
outside
1 - 156
Population Genetic Test Statistics
Pi
149.063481
Theta
140.315842
Tajima's D
0
CLR
0.428454
CSRT
0.368481575921204
Interpretation
Uncertain
