Gene
KWMTBOMO09516 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013049
Annotation
hemolymph_proteinase_19_[Manduca_sexta]
Location in the cell
Extracellular Reliability : 3.047
Sequence
CDS
ATGCTTGGCTTTGTGACCGTTGTGCTGGTTTTAGCACTATCAGTTCGCACACAGGACCAAAGCACTCCAATCTCACCATGCCCGAATGTTTTCGAGTATGAAACGCCGGGGTCAGAGGCTGGGAGGTGGTATGGCGTAGTCCATGTCTCCACTGACAGCACGTTATATTCTCTATGGCTCAATATCGTTCTGGATAGTAAAGCTGATATACTTGGGAATTGGATAGGCGATGTCACAACGCAAGACAACATGAACTTCAAAATCGAAAGCACGCAGACGAAGATCAATCCTGGGCCAGCAACGGCAGTTAGATTTTTCGTCCAATACAATACTTTGAACAAAGCTCCTCGTCTTTTAGCCATCAGATTGAATGGACGTGAAATTTGCAATGCAAACAACCCCCAGCCTGCCCTTGAGAGCCCTCTGTCTTCGGGCGATTCAAAGCCGAACCGATTCGTCACAGCTTCGACTGGTAGACCCCAAACACAACCCAATCGAAGACAACAAACTACTCCATCCCCTAGACCAACAAGTAGACCCTTGAATACTGACTCGAGGACGCCAGAAACTGAAAGACCGCAAATTCCTATTAGAAGACCTCAGGATCCAGTTGTGAGGCCACAGATACCGGATACGAGAACACAGAGCCCTGATGTTCGTCCAGAGCACCTGTTGCAGACTAGACCCGTCCAAGGCCGACCGTCTACCGAAGCCGAGCGACCAGTTTACGTAAAACCCGTGAATTCCGCAAATCCATCGAGCATTTCGGGTGGAGAGGTATCTTCTCAGTCGTATAATCAGAATCAGGACAGAGGGCCTTCGAACAACAATCCTAGGTTCACTCCTGCGCCGCCCACGTACCGTCCACCCTCGAAACAGCCTAGCTCATCACAACACCACGACGACAATGGAAGTGCAGGAGACGTTGACGACTACGAATATTTCACGGGAGGCCAGCCCACCATCATATTGCCGAGCAGCAACCGAAGCGATCAGAATACCTGCGGCCGAGTAATGTTGAACAGGCCTATACCTTTAGTTCTGAACGGGGTCCCGACGTTAGAAGGTCAATGGCCCTGGCAGATCGCCTTATACCAGACTCAGAGAGTAGACAACAAATACATATGCGGAGGAACGCTTGTGTCACACCGCCACATCATCACGGCGGCACATTGCGTCACAAGGAAGGGCTCTCAACGACTCGTGAACAAGGACACGCTGACCGTGTACTTGGGGAAACACAATCTCAGAACATCCGTCGACGGCGTGCAGATCAGATTCGTCAGTCAAATAATAATACACCCAGAATACAACGCGTCGACATTCAGTCGTGACGTCAGCATCTTGGAGCTGAAAGACAGAGTCACCTATTCGAAATGGGTTCAGCCAGCTTGTCTTTGGCCCAGCAATCAGATCACACTCAATAATATTGTAGGGAAGAAGGGATCAGTGGTAGGCTGGGGCTTTGATGAGACAGGAGTGGCGACAGAAGAGCTCAGCCTGGTGGAGATGCCCGTGGTTGACACTGAAACCTGCATCAGGTCTTACAGCGAATTCTTCGTCAGATTTACATCGGAGTACACTTACTGCGCCGGATATAGAGATGGAACATCGGTCTGTAACGGCGACAGTGGTGGCGGAATGGTCTTCAAGTTTGGCGAGAGCTGGTACCTCAGGGGCCTGGTGTCGCTCTCCGTGGCCAGGCAGAACGAATGCAGATGTGACCCCAGCCATTACGTAGTGTTCAGCGACTTGGCTAAATTCCTACCTTGGCTTCAAGAAAATCTCTTTGACTATTAA
Protein
MLGFVTVVLVLALSVRTQDQSTPISPCPNVFEYETPGSEAGRWYGVVHVSTDSTLYSLWLNIVLDSKADILGNWIGDVTTQDNMNFKIESTQTKINPGPATAVRFFVQYNTLNKAPRLLAIRLNGREICNANNPQPALESPLSSGDSKPNRFVTASTGRPQTQPNRRQQTTPSPRPTSRPLNTDSRTPETERPQIPIRRPQDPVVRPQIPDTRTQSPDVRPEHLLQTRPVQGRPSTEAERPVYVKPVNSANPSSISGGEVSSQSYNQNQDRGPSNNNPRFTPAPPTYRPPSKQPSSSQHHDDNGSAGDVDDYEYFTGGQPTIILPSSNRSDQNTCGRVMLNRPIPLVLNGVPTLEGQWPWQIALYQTQRVDNKYICGGTLVSHRHIITAAHCVTRKGSQRLVNKDTLTVYLGKHNLRTSVDGVQIRFVSQIIIHPEYNASTFSRDVSILELKDRVTYSKWVQPACLWPSNQITLNNIVGKKGSVVGWGFDETGVATEELSLVEMPVVDTETCIRSYSEFFVRFTSEYTYCAGYRDGTSVCNGDSGGGMVFKFGESWYLRGLVSLSVARQNECRCDPSHYVVFSDLAKFLPWLQENLFDY
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
EMBL
Proteomes
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
PDB
6O1G
E-value=4.50948e-28,
Score=312
Ontologies
GO
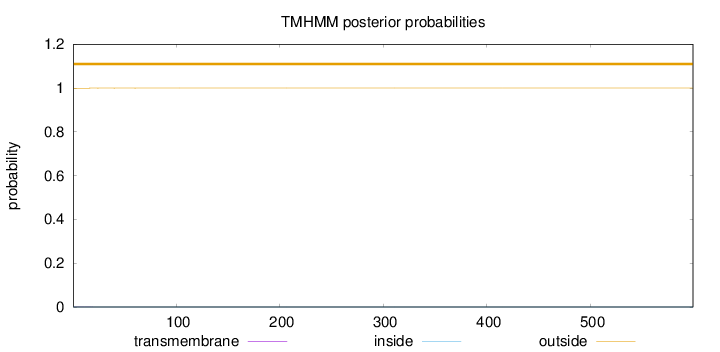
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.994599
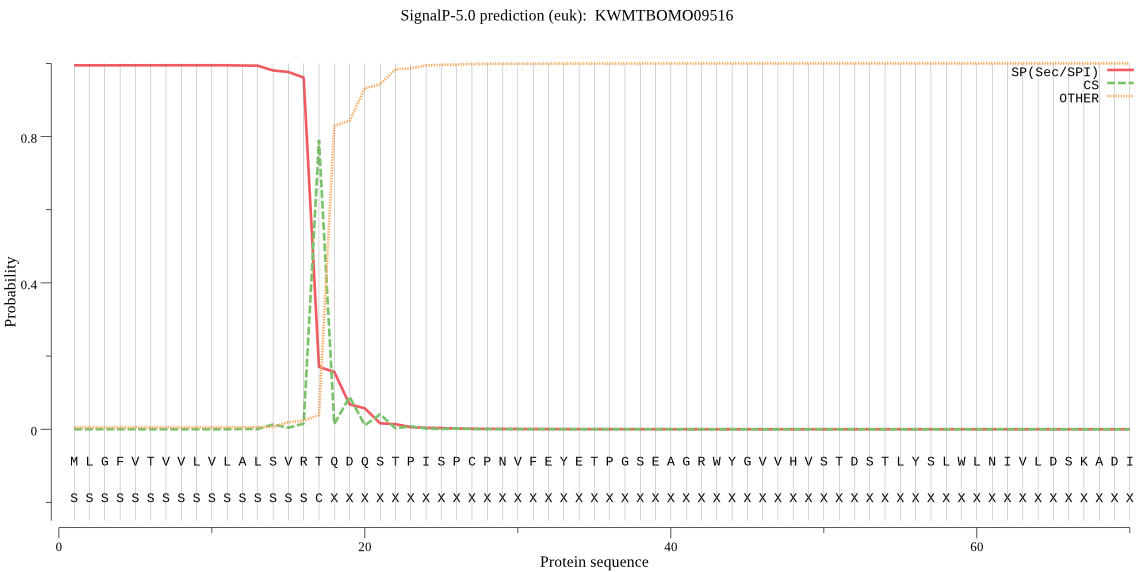
Length:
599
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00568999999999998
Exp number, first 60 AAs:
0.00505
Total prob of N-in:
0.00040
outside
1 - 599
Population Genetic Test Statistics
Pi
191.893781
Theta
168.438066
Tajima's D
0.361113
CLR
0.231338
CSRT
0.469026548672566
Interpretation
Uncertain
