Gene
KWMTBOMO09515
Annotation
olfactory_receptor_57_[Bombyx_mori]
Full name
Odorant receptor
Location in the cell
PlasmaMembrane Reliability : 2.682
Sequence
CDS
ATGAACAAGAGGAATCTGATCACATCAGACGAAGAAAAAGTCATTACAGAAAGGCTTCGTAATATAATTTCGCAGCATAAGCGCGCTTTGGACGCAGCAGAGGCTATCAAGCATTACTTATCCGGAGCTCTGTTGGTACAACTCATGGTGTCCATAGTAGTCATCTGTACTACGGCATATCAACTGGCTGTGAAAAAATCAACTACTATGCAATCGCTGACAATGGCTGGATATCTATTTGGAACATCATTGGAAGTCTTCTTATTTTGTTATCAAGGGGAATTCCTTCGTGAATCTAGTGAAGAAATCGCAGACGCCGCATATGAATGTCCATGGTACACTCTCACTCGACCCTTGAAAAAGACACTGTTAATAATCATGACGCGAGCACAGAGACCCGCCACACTCACGGCAGGTGGATTCGTGACTTTGGACATAACCGAATATATGGCGATAATGAAAGCATCCTACAGCTTTTTCACGGTTCTTCAACAAGTCAGCGAATAA
Protein
MNKRNLITSDEEKVITERLRNIISQHKRALDAAEAIKHYLSGALLVQLMVSIVVICTTAYQLAVKKSTTMQSLTMAGYLFGTSLEVFLFCYQGEFLRESSEEIADAAYECPWYTLTRPLKKTLLIIMTRAQRPATLTAGGFVTLDITEYMAIMKASYSFFTVLQQVSE
Summary
Similarity
Belongs to the insect chemoreceptor superfamily. Heteromeric odorant receptor channel (TC 1.A.69) family.
Uniprot
C7SFP3
C4B7Y5
A7E3I7
C7SFP2
B1B1Q3
F7VJP4
+ More
C4B7X5 A7E3I9 A2T876 C4B7X6 A7E3I8 A0A0P1IVK0 B1B1Q9 A7E3I6 A0A097ITV0 A0A0B5CST1 A0A097IYP3 A0A1B3P5P0 A0A0U3C2H9 C4B7Y6 A0A0B5A1I7 A0A0K8TU69 A0A2K8GKR4 A0A2H1VP78 A0A075TD87 A0A2A4J5G3 A0A076E9B0 A0A1X9P7L2 A0A076E5Y2 A0A1B3B762 A0A2L1TG71 A0A076E7N6 A0A0S1TPX9 A0A2W1BIJ7 A0A2H1VP84 A0A2K8GKU0 A0A1Y9TJT0 A0A3G6V6Q7 A0A3S2LFZ6 A2T875 A0A1B3P5R4 A0A076E9A1 A0A0U3AAG9 A0A2W1BT33 H9JXG3 A0A0E4FLW9 A0A0E4FJN0 A0A076E9C9 A0A2K8GKT1 A0A345BEY7 A0A2K8GL58 A0A1S5XXQ2 A0A386H924 A0A1S5XXP9 A0A386H9H2 C4B7Z2 A0A0P1IVI8 A0A194RGK4 A0A0N1I9U0 A0A3Q8HNT4 A0A075T6X9 H9A5N4 A0A076E9A5 A0A1W6L1A9 A0A3L9M106 A0A084VRJ3 R9PST7 A0A0S3J2U6 A0A310S9J6 A0A2P1JHD8 A0A0S3J2Q0 A0A158P0M9 Q0IGE1 A0A1W6L1H0 A0A139WMR1 A0A026WNA7 A0A3L8DXS0 A0A182QFE2 A0A0M4IUD6 A0A158P0M5 A0A0E3Y6Y9 A0A221I0D8 A0A2J7Q6A5 A0A182WR07 A0A1W6L1G3 A0A182S4P8 A0A3S5HSS8 A0A221I0H5 A0A3T0CIR4 E2A3X6 A0A0C5DMP0 A0A182PUY6 A0A0C5DAR1 A0A0E3U2J9 A0A182M592 A0A139WMB1 A0A0P1IW06 A0A1J0KKH4 D6WCW3
C4B7X5 A7E3I9 A2T876 C4B7X6 A7E3I8 A0A0P1IVK0 B1B1Q9 A7E3I6 A0A097ITV0 A0A0B5CST1 A0A097IYP3 A0A1B3P5P0 A0A0U3C2H9 C4B7Y6 A0A0B5A1I7 A0A0K8TU69 A0A2K8GKR4 A0A2H1VP78 A0A075TD87 A0A2A4J5G3 A0A076E9B0 A0A1X9P7L2 A0A076E5Y2 A0A1B3B762 A0A2L1TG71 A0A076E7N6 A0A0S1TPX9 A0A2W1BIJ7 A0A2H1VP84 A0A2K8GKU0 A0A1Y9TJT0 A0A3G6V6Q7 A0A3S2LFZ6 A2T875 A0A1B3P5R4 A0A076E9A1 A0A0U3AAG9 A0A2W1BT33 H9JXG3 A0A0E4FLW9 A0A0E4FJN0 A0A076E9C9 A0A2K8GKT1 A0A345BEY7 A0A2K8GL58 A0A1S5XXQ2 A0A386H924 A0A1S5XXP9 A0A386H9H2 C4B7Z2 A0A0P1IVI8 A0A194RGK4 A0A0N1I9U0 A0A3Q8HNT4 A0A075T6X9 H9A5N4 A0A076E9A5 A0A1W6L1A9 A0A3L9M106 A0A084VRJ3 R9PST7 A0A0S3J2U6 A0A310S9J6 A0A2P1JHD8 A0A0S3J2Q0 A0A158P0M9 Q0IGE1 A0A1W6L1H0 A0A139WMR1 A0A026WNA7 A0A3L8DXS0 A0A182QFE2 A0A0M4IUD6 A0A158P0M5 A0A0E3Y6Y9 A0A221I0D8 A0A2J7Q6A5 A0A182WR07 A0A1W6L1G3 A0A182S4P8 A0A3S5HSS8 A0A221I0H5 A0A3T0CIR4 E2A3X6 A0A0C5DMP0 A0A182PUY6 A0A0C5DAR1 A0A0E3U2J9 A0A182M592 A0A139WMB1 A0A0P1IW06 A0A1J0KKH4 D6WCW3
Pubmed
19427209
17257213
19100833
25252791
27538507
26657286
+ More
26017144 28736530 25027790 28756777 24998398 26812239 30465940 19121390 29727827 29375398 23950722 26354079 22363688 27006164 24438588 23517120 26626891 29497384 21347285 17635615 17510324 18362917 19820115 24508170 30249741 26265180 25856077 30542294 20798317 25665775 27836740
26017144 28736530 25027790 28756777 24998398 26812239 30465940 19121390 29727827 29375398 23950722 26354079 22363688 27006164 24438588 23517120 26626891 29497384 21347285 17635615 17510324 18362917 19820115 24508170 30249741 26265180 25856077 30542294 20798317 25665775 27836740
EMBL
FJ170766
ACH73301.1
AB472134
BAH66351.1
BK005952
DAA06002.1
+ More
FJ170765 ACH73300.1 AB186512 BAG12811.1 BK006738 DAA34890.1 AB472123 BAH66341.1 BK005954 DAA06004.1 DQ991160 ABK27854.1 AB472124 BAH66342.1 BK005953 DAA06003.1 LN885125 CUQ99414.1 AB186518 AB472122 BAG12817.1 BAH66340.1 BK005951 DAA06001.1 KM597218 AIT69895.1 KM678318 AJE25891.1 KM655556 AIT72006.1 KX655978 AOG12927.1 KP975149 ALT31668.1 AB472135 BAH66352.1 KJ542693 AJD81577.1 GCVX01000203 JAI18027.1 KX585335 ARO70228.1 ODYU01003636 SOQ42639.1 KF768712 KZ150021 AIG51906.1 PZC74884.1 NWSH01002961 PCG67211.1 KF487686 AII01084.1 KX084487 ARO76442.1 KF487663 AII01061.1 KT588148 AOE48058.1 KY707221 AVF19663.1 KF487709 AII01107.1 KR935754 ALM26246.1 PZC74882.1 SOQ42640.1 KX585341 ARO70234.1 KX084481 ARO76436.1 MH193981 AZB49427.1 RSAL01000147 RVE45893.1 DQ991159 ABK27853.1 KX655991 AOG12940.1 KF487670 AII01068.1 KP975153 ALT31672.1 PZC74883.1 BABH01032810 LC002731 BAR43479.1 LC002725 BAR43473.1 KF487705 AII01103.1 KX585340 ARO70233.1 MG816626 AXF48811.1 KY225515 ARO70527.1 KY399302 AQQ73533.1 MG546609 AYD42232.1 KY399301 AQQ73532.1 MG546647 AYD42270.1 AB472140 BAH66358.1 LN885110 CUQ99399.1 KQ460211 KPJ16574.1 KQ460480 KPJ14347.1 MF625588 AXY83415.1 KF768695 AIG51889.1 JN836684 AFC91724.1 KF487675 AII01073.1 KX609510 ARN17917.1 KB466665 RLZ02263.1 ATLV01015691 KE525027 KFB40587.1 GABX01000015 JAA74504.1 KT381575 ALR72581.1 GGOB01000099 MCH29495.1 MG766190 AVN97860.1 KT381548 ALR72554.1 ADTU01005367 BK006160 CH477192 DAA80375.1 EAT48597.1 KX609507 KB465818 ARN17914.1 RLZ02187.1 KQ971312 KYB29105.1 KK107144 EZA57552.1 QOIP01000003 RLU25264.1 AXCN02001476 KP843247 ALD51383.1 ADTU01005364 ADTU01005365 KM251674 AKC58555.1 KY964957 ASM47110.1 NEVH01017534 PNF24115.1 KX609513 KB465925 ARN17920.1 RLZ02205.1 MK048991 AZQ24932.1 KY964940 ASM47093.1 MH050669 AZT78928.1 GL436519 EFN71828.1 KP296771 AJO62235.1 KP296764 AJO62228.1 KM251666 AKC58547.1 AXCM01000144 KQ971317 KYB28981.1 LN885103 CUQ99392.1 KX290655 APC94226.1 KQ971311 EEZ99415.2
FJ170765 ACH73300.1 AB186512 BAG12811.1 BK006738 DAA34890.1 AB472123 BAH66341.1 BK005954 DAA06004.1 DQ991160 ABK27854.1 AB472124 BAH66342.1 BK005953 DAA06003.1 LN885125 CUQ99414.1 AB186518 AB472122 BAG12817.1 BAH66340.1 BK005951 DAA06001.1 KM597218 AIT69895.1 KM678318 AJE25891.1 KM655556 AIT72006.1 KX655978 AOG12927.1 KP975149 ALT31668.1 AB472135 BAH66352.1 KJ542693 AJD81577.1 GCVX01000203 JAI18027.1 KX585335 ARO70228.1 ODYU01003636 SOQ42639.1 KF768712 KZ150021 AIG51906.1 PZC74884.1 NWSH01002961 PCG67211.1 KF487686 AII01084.1 KX084487 ARO76442.1 KF487663 AII01061.1 KT588148 AOE48058.1 KY707221 AVF19663.1 KF487709 AII01107.1 KR935754 ALM26246.1 PZC74882.1 SOQ42640.1 KX585341 ARO70234.1 KX084481 ARO76436.1 MH193981 AZB49427.1 RSAL01000147 RVE45893.1 DQ991159 ABK27853.1 KX655991 AOG12940.1 KF487670 AII01068.1 KP975153 ALT31672.1 PZC74883.1 BABH01032810 LC002731 BAR43479.1 LC002725 BAR43473.1 KF487705 AII01103.1 KX585340 ARO70233.1 MG816626 AXF48811.1 KY225515 ARO70527.1 KY399302 AQQ73533.1 MG546609 AYD42232.1 KY399301 AQQ73532.1 MG546647 AYD42270.1 AB472140 BAH66358.1 LN885110 CUQ99399.1 KQ460211 KPJ16574.1 KQ460480 KPJ14347.1 MF625588 AXY83415.1 KF768695 AIG51889.1 JN836684 AFC91724.1 KF487675 AII01073.1 KX609510 ARN17917.1 KB466665 RLZ02263.1 ATLV01015691 KE525027 KFB40587.1 GABX01000015 JAA74504.1 KT381575 ALR72581.1 GGOB01000099 MCH29495.1 MG766190 AVN97860.1 KT381548 ALR72554.1 ADTU01005367 BK006160 CH477192 DAA80375.1 EAT48597.1 KX609507 KB465818 ARN17914.1 RLZ02187.1 KQ971312 KYB29105.1 KK107144 EZA57552.1 QOIP01000003 RLU25264.1 AXCN02001476 KP843247 ALD51383.1 ADTU01005364 ADTU01005365 KM251674 AKC58555.1 KY964957 ASM47110.1 NEVH01017534 PNF24115.1 KX609513 KB465925 ARN17920.1 RLZ02205.1 MK048991 AZQ24932.1 KY964940 ASM47093.1 MH050669 AZT78928.1 GL436519 EFN71828.1 KP296771 AJO62235.1 KP296764 AJO62228.1 KM251666 AKC58547.1 AXCM01000144 KQ971317 KYB28981.1 LN885103 CUQ99392.1 KX290655 APC94226.1 KQ971311 EEZ99415.2
Proteomes
PRIDE
Pfam
PF02949 7tm_6
Interpro
IPR004117
7tm6_olfct_rcpt
ProteinModelPortal
C7SFP3
C4B7Y5
A7E3I7
C7SFP2
B1B1Q3
F7VJP4
+ More
C4B7X5 A7E3I9 A2T876 C4B7X6 A7E3I8 A0A0P1IVK0 B1B1Q9 A7E3I6 A0A097ITV0 A0A0B5CST1 A0A097IYP3 A0A1B3P5P0 A0A0U3C2H9 C4B7Y6 A0A0B5A1I7 A0A0K8TU69 A0A2K8GKR4 A0A2H1VP78 A0A075TD87 A0A2A4J5G3 A0A076E9B0 A0A1X9P7L2 A0A076E5Y2 A0A1B3B762 A0A2L1TG71 A0A076E7N6 A0A0S1TPX9 A0A2W1BIJ7 A0A2H1VP84 A0A2K8GKU0 A0A1Y9TJT0 A0A3G6V6Q7 A0A3S2LFZ6 A2T875 A0A1B3P5R4 A0A076E9A1 A0A0U3AAG9 A0A2W1BT33 H9JXG3 A0A0E4FLW9 A0A0E4FJN0 A0A076E9C9 A0A2K8GKT1 A0A345BEY7 A0A2K8GL58 A0A1S5XXQ2 A0A386H924 A0A1S5XXP9 A0A386H9H2 C4B7Z2 A0A0P1IVI8 A0A194RGK4 A0A0N1I9U0 A0A3Q8HNT4 A0A075T6X9 H9A5N4 A0A076E9A5 A0A1W6L1A9 A0A3L9M106 A0A084VRJ3 R9PST7 A0A0S3J2U6 A0A310S9J6 A0A2P1JHD8 A0A0S3J2Q0 A0A158P0M9 Q0IGE1 A0A1W6L1H0 A0A139WMR1 A0A026WNA7 A0A3L8DXS0 A0A182QFE2 A0A0M4IUD6 A0A158P0M5 A0A0E3Y6Y9 A0A221I0D8 A0A2J7Q6A5 A0A182WR07 A0A1W6L1G3 A0A182S4P8 A0A3S5HSS8 A0A221I0H5 A0A3T0CIR4 E2A3X6 A0A0C5DMP0 A0A182PUY6 A0A0C5DAR1 A0A0E3U2J9 A0A182M592 A0A139WMB1 A0A0P1IW06 A0A1J0KKH4 D6WCW3
C4B7X5 A7E3I9 A2T876 C4B7X6 A7E3I8 A0A0P1IVK0 B1B1Q9 A7E3I6 A0A097ITV0 A0A0B5CST1 A0A097IYP3 A0A1B3P5P0 A0A0U3C2H9 C4B7Y6 A0A0B5A1I7 A0A0K8TU69 A0A2K8GKR4 A0A2H1VP78 A0A075TD87 A0A2A4J5G3 A0A076E9B0 A0A1X9P7L2 A0A076E5Y2 A0A1B3B762 A0A2L1TG71 A0A076E7N6 A0A0S1TPX9 A0A2W1BIJ7 A0A2H1VP84 A0A2K8GKU0 A0A1Y9TJT0 A0A3G6V6Q7 A0A3S2LFZ6 A2T875 A0A1B3P5R4 A0A076E9A1 A0A0U3AAG9 A0A2W1BT33 H9JXG3 A0A0E4FLW9 A0A0E4FJN0 A0A076E9C9 A0A2K8GKT1 A0A345BEY7 A0A2K8GL58 A0A1S5XXQ2 A0A386H924 A0A1S5XXP9 A0A386H9H2 C4B7Z2 A0A0P1IVI8 A0A194RGK4 A0A0N1I9U0 A0A3Q8HNT4 A0A075T6X9 H9A5N4 A0A076E9A5 A0A1W6L1A9 A0A3L9M106 A0A084VRJ3 R9PST7 A0A0S3J2U6 A0A310S9J6 A0A2P1JHD8 A0A0S3J2Q0 A0A158P0M9 Q0IGE1 A0A1W6L1H0 A0A139WMR1 A0A026WNA7 A0A3L8DXS0 A0A182QFE2 A0A0M4IUD6 A0A158P0M5 A0A0E3Y6Y9 A0A221I0D8 A0A2J7Q6A5 A0A182WR07 A0A1W6L1G3 A0A182S4P8 A0A3S5HSS8 A0A221I0H5 A0A3T0CIR4 E2A3X6 A0A0C5DMP0 A0A182PUY6 A0A0C5DAR1 A0A0E3U2J9 A0A182M592 A0A139WMB1 A0A0P1IW06 A0A1J0KKH4 D6WCW3
PDB
6C70
E-value=2.37526e-12,
Score=169
Ontologies
GO
PANTHER
Topology
Subcellular location
Cell membrane
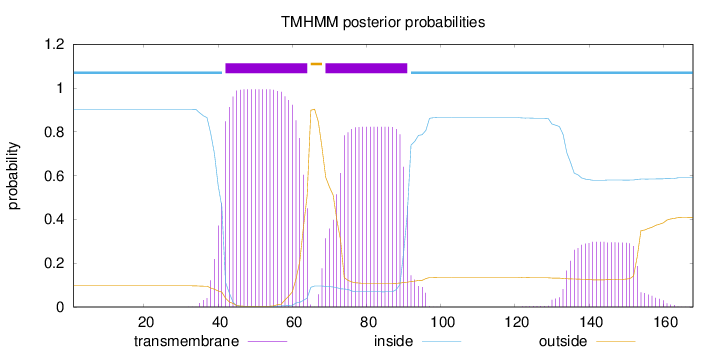
Length:
168
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
45.6085800000001
Exp number, first 60 AAs:
19.72094
Total prob of N-in:
0.90336
POSSIBLE N-term signal
sequence
inside
1 - 41
TMhelix
42 - 64
outside
65 - 68
TMhelix
69 - 91
inside
92 - 168
Population Genetic Test Statistics
Pi
145.884712
Theta
199.318744
Tajima's D
-1.07732
CLR
217.165902
CSRT
0.12324383780811
Interpretation
Uncertain
