Gene
KWMTBOMO09514
Annotation
PREDICTED:_uncharacterized_protein_LOC101747137_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 1.533
Sequence
CDS
ATGGCGCAGGCGATAGCGGACGAATTGGACATCGCAGTCGGTGTAAGGACGTACTGGACTGACTCCAGTACAGTCCTGACATGGATAAAGACCGACCCACGCACGTTCAAACCTTTCGTCGCGCACCGACTCGCCGAGATAGAAGAGTCAACGAAGCCCCAAGAATGGCGATGGGTGCCTGGCTCGCAAAACCCAGCAGACGACGCAACTAGAGAGGCGCCGGCGGATTTCGACCACACGCATCGGTGGTTTAATGGACCCGAGTTCCTGACGTGGGATGAATCGCGCTGGCCCAAGCCGCGAACATTCAAGCAAGAACCGTCTGGAGAAGAGAAAGAGGCCTACCTGGTCGCTACGGCGAGGACCGCTGAAGCTCAACCCACGCCCGATCCGCACAGATTCTCGAGCTGGGTCAGGTTATTGAGAGCAACAGCCAGGGTCCTCCAATTTATCGAGCTGTGTCGACCCCGGAAGGAGAGCGCCTGCGTGTCCAGACAATCGAAGCAGCAAGATCCCACGTGGAGGACGACACGAACGAAGCAACCCCGATCCACATGGAAGTTAAGGACCCCGGAAGCACCTACAGAAGCATGGCTGCCGCTTGATCCGCCCCACTTGAAGAAGGCGGAGAAGATCCTACTGAGAAACAGCCAAGGAGAGAGCTTCGGTGAAAAAGATCCCGAACGTCACCCCAAGTTGCGACGGCTCGACGTCGTGATGGAAGATGGCCTGTTGCGCCTGCGCGGACGTATCGACGCAGCGCAATACATAGATGCAGGCTGCAAGCGACCGATCGTGCTGGACGGGAAGCACGTGATAGCGAGATTATTGATCAAACACTATCACGAGGCGTTCCAGCACGGTAACCACGCAACGGTGATGAACGAGTCATTTTTCGATGTGGCCATGGCGTTCTTTATAAAAAACAAAATGTTTGGTTTAACAATAACTTTGAACACTTTATCATGGGCTGGATTAATAATGCGAGATCAATACACGAAGACTCAAAGGATCATCGTCAGAGTGTATGCATGGCTGGTTTTCCTATATCTATTTGTTGCTGCAGCTTGCGTACAAATCGCGGACATTATTGACATCTGGGGTGACATTAATCTCATGGCGGAAACAGCTTTGCTTCTCTTTATGGAATTCGCAGTGATTTCTAAGATTTTAACCTTGTTATTGAGATACGATAGAATAATGGAGATCATCAATGGAACTGAAGAAATATTGTATTTTGAAAACGGATTGGAAGGCCAAAGGATTATCGCAAGGATGGTTCTGTCCAATGGTTCTGTAAAGGCTTGGCTTGGCTTGGCCATGGCTTTGGTAAAGCTGTCGTTTGCTAATACCGAAATCAATTACAATAATTAG
Protein
MAQAIADELDIAVGVRTYWTDSSTVLTWIKTDPRTFKPFVAHRLAEIEESTKPQEWRWVPGSQNPADDATREAPADFDHTHRWFNGPEFLTWDESRWPKPRTFKQEPSGEEKEAYLVATARTAEAQPTPDPHRFSSWVRLLRATARVLQFIELCRPRKESACVSRQSKQQDPTWRTTRTKQPRSTWKLRTPEAPTEAWLPLDPPHLKKAEKILLRNSQGESFGEKDPERHPKLRRLDVVMEDGLLRLRGRIDAAQYIDAGCKRPIVLDGKHVIARLLIKHYHEAFQHGNHATVMNESFFDVAMAFFIKNKMFGLTITLNTLSWAGLIMRDQYTKTQRIIVRVYAWLVFLYLFVAAACVQIADIIDIWGDINLMAETALLLFMEFAVISKILTLLLRYDRIMEIINGTEEILYFENGLEGQRIIARMVLSNGSVKAWLGLAMALVKLSFANTEINYNN
Summary
Uniprot
EMBL
KQ459986
KPJ18861.1
LNIX01000019
OXA44855.1
LNIX01000015
OXA46575.1
+ More
LNIX01000039 OXA39294.1 AAZX01023278 JXUM01013493 KQ560378 KXJ82704.1 JXUM01127084 KQ567147 KXJ69602.1 GEZM01052587 JAV74399.1 GEZM01052586 JAV74401.1 GEHC01000913 JAV46732.1 GGFJ01001476 MBW50617.1 GGFJ01001462 MBW50603.1 JXUM01009840 KQ560293 KXJ83223.1 JXUM01130316 KQ567562 KXJ69370.1
LNIX01000039 OXA39294.1 AAZX01023278 JXUM01013493 KQ560378 KXJ82704.1 JXUM01127084 KQ567147 KXJ69602.1 GEZM01052587 JAV74399.1 GEZM01052586 JAV74401.1 GEHC01000913 JAV46732.1 GGFJ01001476 MBW50617.1 GGFJ01001462 MBW50603.1 JXUM01009840 KQ560293 KXJ83223.1 JXUM01130316 KQ567562 KXJ69370.1
Proteomes
Pfam
Interpro
IPR036397
RNaseH_sf
+ More
IPR041588 Integrase_H2C2
IPR040676 DUF5641
IPR008737 Peptidase_asp_put
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR001995 Peptidase_A2_cat
IPR001584 Integrase_cat-core
IPR001878 Znf_CCHC
IPR006094 Oxid_FAD_bind_N
IPR016166 FAD-bd_PCMH
IPR016164 FAD-linked_Oxase-like_C
IPR005312 DUF1759
IPR016170 Cytok_DH_C_sf
IPR015213 Cholesterol_OX_subst-bd
IPR036318 FAD-bd_PCMH-like_sf
IPR016169 FAD-bd_PCMH_sub2
IPR016167 FAD-bd_PCMH_sub1
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR041588 Integrase_H2C2
IPR040676 DUF5641
IPR008737 Peptidase_asp_put
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR001995 Peptidase_A2_cat
IPR001584 Integrase_cat-core
IPR001878 Znf_CCHC
IPR006094 Oxid_FAD_bind_N
IPR016166 FAD-bd_PCMH
IPR016164 FAD-linked_Oxase-like_C
IPR005312 DUF1759
IPR016170 Cytok_DH_C_sf
IPR015213 Cholesterol_OX_subst-bd
IPR036318 FAD-bd_PCMH-like_sf
IPR016169 FAD-bd_PCMH_sub2
IPR016167 FAD-bd_PCMH_sub1
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
ProteinModelPortal
Ontologies
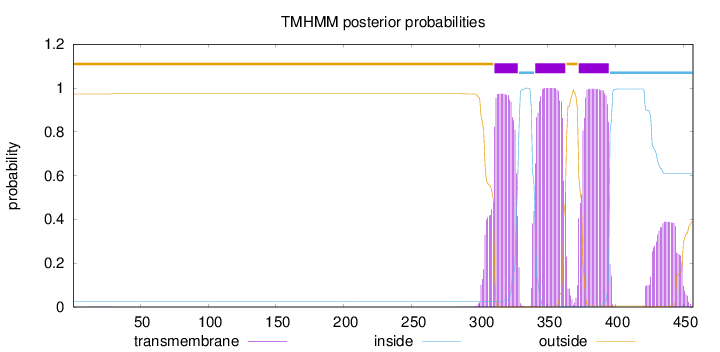
Topology
Length:
457
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
71.60632
Exp number, first 60 AAs:
0.01384
Total prob of N-in:
0.02671
outside
1 - 310
TMhelix
311 - 328
inside
329 - 340
TMhelix
341 - 363
outside
364 - 372
TMhelix
373 - 395
inside
396 - 457
Population Genetic Test Statistics
Pi
361.682861
Theta
212.496478
Tajima's D
1.647029
CLR
176.409172
CSRT
0.821908904554772
Interpretation
Uncertain
